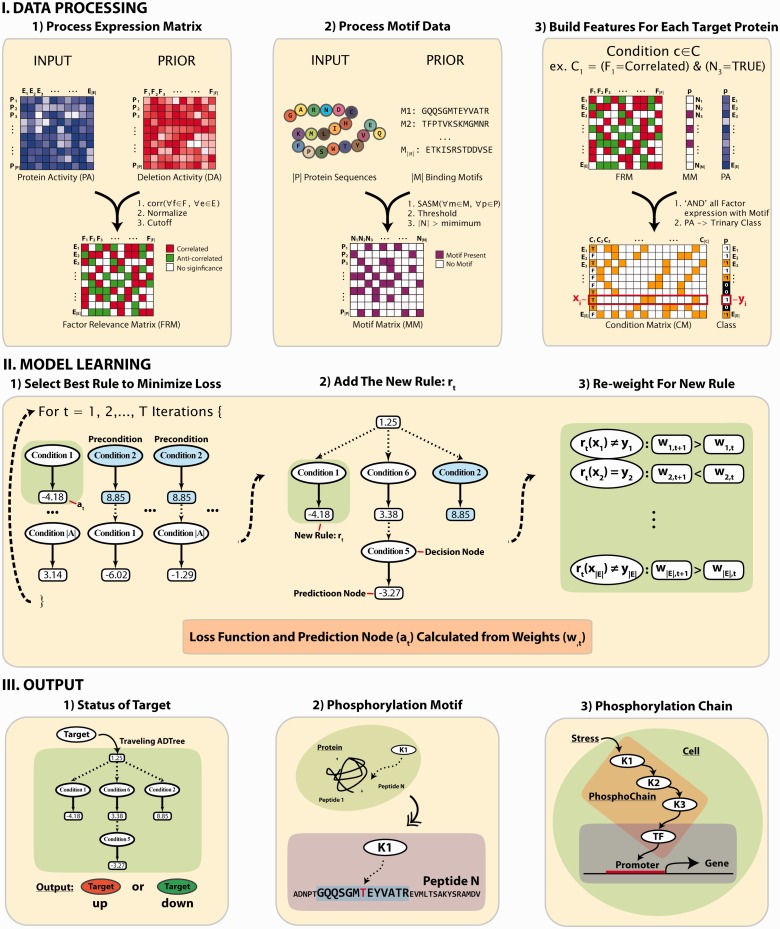
Fig. 1.
Overall strategy to identify phosphorylation network for PhosphoChain. DATA PROCESSING: PhosphoChain constructs three matrices from the input data to generate the model: an FRM, a binding MM and a CM. FRM is calculated using the PCC between the input PA profile and factor DA profile. MM is generated from a list of template peptide motifs calculated based on the similarity to known phosphorylation motifs as measured by the structure-derived amino acid substitution matrix, SASM. CM is generated by combining the FRM and MM. MODEL LEARNING: For each target protein, an ADTree composed of decision nodes and prediction nodes is generated from the CM. Decision nodes specify the predicate condition that (i) the kinase/phosphatase has a high correlation/anti-correlation with the experiment; and (ii) the phosphorylated motif is present on the protein sequence. Each prediction node contains a score associated with a decision node. By traversing all satisfied nodes on the ADTree, target activity is predicted. OUTPUT: The PhosphoChain model predicts (i) target activity; (ii) phosphorylation sites; and (iii) protein phosphorylation chains