Abstract
The metabolic enzyme for folate, Aldh1L1, has been shown to be expressed robustly in astrocytes of the brain. It is now well accepted that astrocytes in certain regions of the adult brain also serve as neural stem cells. Here, we examined whether Aldh1L1 is also expressed in postnatal neural stem cells. In vitro, cells in neural stem cell culture conditions have robust Aldh1L1 promoter activity. In vivo, in the adult brain, astroctyes in neurogenic regions express Aldh1L1 in a pattern consistent with inclusion in neural stem cells, and analysis of Aldh1L1+ cell transcriptome profiles from neurogenic regions reveals a robust enrichment of known regulators of neurogenesis. Genetic fate mapping with Aldh1L1 BAC Cre animals reveals adult born neuroblasts of the rostral migratory stream are derived from Aldh1L1 expressing cells, as are sporadic neurons in other regions of the brain. Combining these lines of evidence from transgenic animals, cell culture, transcriptome profiling, and fate mapping, we conclude that Aldh1L1 is also expressed in neural stem cells in the brain. These findings may influence the future design of experiments utilizing Aldh1L1 genetic tools, and also suggest existing Aldh1L1 bacTRAP mice may be of use for further experiments profiling neural stem cells in vivo.
Introduction
The astrocyte is a markedly important cell for the normal functioning of the CNS: they provide essential metabolic support for neurons, key signals for synaptogenesis(Christopherson et al. 2005; Eroglu and Barres 2010; Ullian et al. 2001), and actively respond to neurotransmission(Sun et al. 2013). Classically, astrocytes were most commonly identified by staining with the marker Glial Fibrillary Acidic Protein (Gfap), however this particular antigen readily labels astrocytes in only a subset of brain regions, or during gliosis(Chiu and Goldman 1985; Eng et al. 2000). Recently new astrocyte markers, notably the Aldehyde dehydrogenase 1 family, member L1 (Aldh1L1) have become popular. By several accounts, Aldh1L1 more robustly labels astrocytes in the brain, including all GFAP+ cells, but also with expression in parenchymal astrocytes poorly labeled by GFAP (Anthony and Heintz 2007; Cahoy et al. 2008; Dougherty et al. 2012b; Doyle et al. 2008; Neymeyer et al. 1997; Pfrieger and Slezak 2012; Yang et al. 2011), and there now exist both reliable antibodies for post hoc labeling(Krupenko and Oleinik 2002; Rhodes and Trimmer 2006), and validated a Bacterial Artificial Chromosome (BAC) for genetic targeting (Anthony and Heintz 2007).
Cells with features of astrocytes serve as neural stem cells in the neurogenic regions of the brain – the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) of the dentate gyrus (Alvarez-Buylla and Garcia-Verdugo 2002; Kempermann 2002). In both regions, slowly dividing astrocytes (Gfap+), give rise to more rapidly cycling neuronal progenitors (transiently amplifying cells), which eventually give rise to cells destined to become neurons to the olfactory bulb or dentate gyrus granule cell layer, respectively (Doetsch 2003; Garcia et al. 2004). These adult stem cells arise from a transient developmental cell type, which can also serve as a neural stem cell, the radial glia (Merkle et al. 2004). Radial glia are also known to express Aldh1L1, largely starting as they decrease their proliferation after the period of neuronal production (Anthony and Heintz 2007). However, there are some remaining questions about these cells, both during development and adulthood.
Previously, we have employed an Aldh1L1 BAC for transgenesis, which reliably and robustly labels astrocytes in the adult brain, targeting these cells for Translating Ribosome Affinity Purification(TRAP) (Doyle et al. 2008; Heiman et al. 2008) and sophisticated multicolored genetic imaging (Dougherty et al. 2012b). Here, we address the question of whether the astrocytes labeled by Aldh1L1 include the neural stem cells and map the fate of cells with Aldh1L1 BAC transcriptional activity. Examination of both the BAC transgene expression, as well as antibody labeling and fate mapping, determines that postnatal neural stem cells, in vivo, express Aldh1L1.
Materials and Methods
Mouse models
All experiments with mice were approved by the animal care and use committee of Rockefeller University We utilized multiple lines generated with the same BAC (RP23-7M9): the TRAP line Aldh1L1 JD130 (Doyle et al. 2008), the Prism lines driving either Cerulean (JD1849) or DSredmax(JD1989) (Dougherty et al. 2012b), and a Cre expressing line (JD1884) (Tien et al. 2012). All BAC transgenic mice were maintained as heterozygotes, and genotyped at each generation by tail tip PCR or fluorescence microscopy. Cre mice were crossed to reporter mice with a floxed stop in front of Yfp, in the Rosa locus (b6.129×1-GT(ROSA)26Sor tm1(EYFP)Cos/J, Jackson Laboratories) for fate mapping experiments.
Neuroanatomy
For immunohistochemistry, brains were processed using MultiBrain™ Technology (NeuroScience Associates) using a 1:75,000 dilution of Goat anti-EGFP serum (Heiman et al. 2008) following the Vectastain elite protocol (Vector Labs). Note: GFP antibodies cross react with the YFP expressed here. Serial sections were digitized with Zeiss Axioskop2 microscope at 10× magnification. For immunofluorescence, sections were blocked with 5% normal donkey serum, 0.25% triton and incubated with primary antibodies (Ng2, NeuN from Chemicon, and NF200 from Sigma, Aldh1L1, a gift from Dr. Krupenko), and appropriate Alexa dye conjugated secondary antibodies (Invitrogen). Prism mouse images were taken with a LSM 510 NLO inverted multiphoton system. For five color imaging, system was utilized as a standard confocal: Yfp was excited with a 514 nm laser, and detected with a Band Pass 540/20 filter in place. mCherry and DSredmax were excited with a 543 nm laser and detected with a 565-615 IP Band Pass filter. Cerulean was exited with a 458 nm laser, as detected with a 470-500 Band Pass filter. Far red Alexa dyes were excited with a 633 nm laser and collected with the Lambda mode of the Zeiss system. DAPI was visualized with a coherent chameleon 2 photon on the same system. Epifluorescence images were taken with a wide-field Nikon microscope.
Microarray analysis
We reanalyzed data from GEO: GSE13379(Doyle et al. 2008) as follows: Cel files for Aldh1L1 TRAP microarray data from forebrain and cerebellum(3 replicates each from, each replicate from pooled adult animals) were normalized with GCMRA using Bioconductor within the statistical package R, and chip definition files based on Entrez Gene Ids(Dai et al. 2005). Data were filtered to remove genes with low expression (less than 50), and to keep only genes enriched in astrocytes(TRAP/Total tissue RNA fold change >1), prior to directly comparing TRAP samples from Aldh1L1 Forebrain and Aldh1L1 Cerebellum utilizing the LIMMA module with Benjamini-Hochberg multiple testing correction. A threshold for forebrain enriched genes was selected at p<.05 with a 2 fold enrichment. pSI was calculated only for forebrain Aldh1L1 as described(Dougherty et al. 2010) comparing this sample to all of our other previously collected TRAP samples(Dalal et al. in press; Dougherty et al. 2013; Doyle et al. 2008). Analyzed data are available in Supplemental Table 1.
Cell culture
Postnatal day six mice of line Prism JD1989 were euthanized, and cortices were dissociated with Trypsin and fire polished pipettes, and seeded in either Neurosphere media (DMEM/F12, 1% Penicillin Streptomycin, 2% B27 Supplement (Invitrogen), 10ng/ml bFGF, 10 ng/ml EGF (New England Biolabs), 5 ug/ml Heparin(Sigma)) or traditional astrocyte media (DMEM/F12, 1% Penicillin Streptomycin, 10% Fetal Bovine Serum(Sigma)), at 50,000 cells per ml. Growth factor was added to neurospheres two times a week, and cells were passaged at one week by dissociation with trypsin on seeding on poly-ornithine fibronectin coated plates as described (Nakano et al. 2005).
Results
Aldh1L1 transcriptional activity in neurosphere cultures
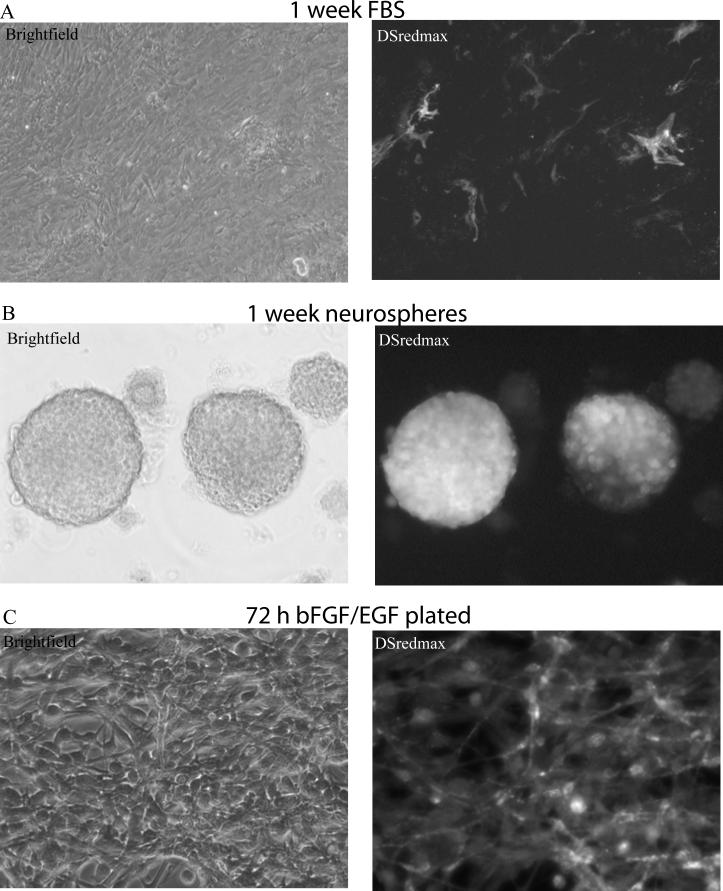
Previously, we generated triple colored transgenic mice, with three BACs in a single genomic locus, to label neurons, astrocytes and oligodendrocytes with distinct fluorophores in vivo (Dougherty et al. 2012b). Here, we tested one of these mouse lines, Prism JD1989, for the same activity in vitro. The three cell types are labeled by BACs covering Aldh1L1(astrocytes, DSredmax), Snap25(neuron, YFP-L10a), and Mobp(oligodendrocytes, Cerulean). From post-natal day 6 mouse cortices, we cultured both neural stem cells, with the classic ‘neurosphere’ technique (Reynolds and Weiss 1992) including the mitogens EGF and bFGF, and also cultured cortical astrocytes, by plating these cells in traditional astrocyte conditions (McCarthy and de Vellis 1980; Nakano et al. 2005). Contrary to our expectations, after one week in vitro, only a fraction of the cells plated under astrocyte conditions demonstrated expression of the DSredmax transgene from the Aldh1L1 BAC promoter (Figure 1A). In contrast, nearly all of the neurospheres (Figure 1B) showed bright DSredmax expression, though some spheres had varying degrees of DSredmax negative cells within them, suggesting some mosaicism.
Figure 1. Aldh1L1 is robustly expressed in neural stem cell cultures.
A) In cultures derived from a P7 mouse cortex from a mouse line, in which an Aldh1L1 BAC promoter drives DSredmax, relatively few cells express DSredmax when plated for one week under standard astrocyte culture conditions. B) In contrast, in neurosphere culture conditions (with EGF and bFGF), many cells robustly express dsredmax. C) Plated cells from these neurospheres continue to show DSredmax expression in the presence of EGF and bFGF
Neurospheres are floating cultures, while traditional astrocyte conditions grow as adherent cultures. To determine if the Aldh1L1 BAC activity was simply suppressed by plating, we dissociated the neurospheres and plated them as neural progenitor cultures, still in the presence of mitogen. Under these conditions the cells sustained bright expression of DSredmax from the Aldh1L1 BAC, indicating that adherent culture conditions do not suppress Aldh1L1 BAC activity (Figure 1C). Likewise, induction of differentiation by withdrawal of mitogen did result in the emergence of transcriptional activity from the Mobp BAC, as evidenced by Cerulean positive cells with oligodendroglial morphology(not shown), but did not substantially decrease DSredmax fluorescence. It is possible that the transgene in vitro was not accurately reflecting the expression of the endogenous gene. To provide a more direct assessment of Aldh1L1 gene expression, we also examined Aldh1L1 expression by RT-qPCR on cells from the same culture conditions but derived from E14.5 wildtype mouse cortex. Aldh1L1 mRNA was robustly detected in the neural stem cell culture conditions, crossing threshold of detection around 21 cycles, not much later than the neural stem cell marker Nestin (19 cycles). Analyzing the data with the ddCT method relative to Actb(Beta Actin) mRNA, expression of Aldh1L1 decreased with differentiation approximately 3 fold. In total, our experiments suggested the Aldh1L1 may be expressed in neural stem cells under a variety of culture conditions.
Aldh1L1 is expressed in neurogenic niches in vivo
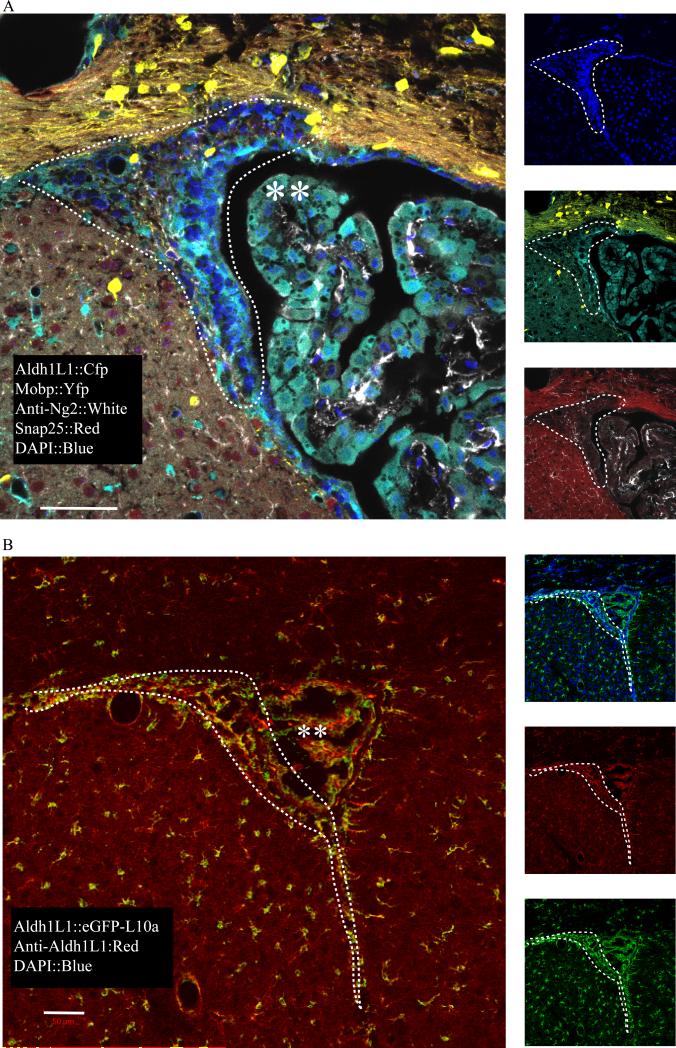
Gene expression in culture does not always reflect the in vivo state. To determine if Aldh1L1 could be expressed in post-natal neural stem cells in vivo we examined its pattern of expression in regions of adult neurogenesis across multiple BAC transgenics and using an antibody to the endogenous protein. In the neurogenic SVZ, Cerulean from an Aldh1L1 BAC (in Prism line JD1849) is found in a pattern consistent with known localization of ‘Type B’ neural stem cells (Doetsch 2003; Dougherty et al. 2005; Nakano et al. 2007): Cerulean+ cells surround pockets of Cerulean−, DAPI+ nuclei(presumptive neuroblasts) (Figure 2B). These Cerulean expressing cells show no overlap with Mobp BAC activity (oligodendrocytes, Yfp), Snap25 BAC activity (mature neurons, mCherry), or Ng2 antibody staining (oligodendrocytes progenitor cells, OPCs). Likewise, in the SGZ, the Aldh1L1 BAC labels the astrocytes found immediately beneath the granule cell layer, consistent with a ‘Type B’ neural stem cell (not shown). We see this neurogenic labeling across all of our Aldh1L1 transgenic lines. For example, in mouse line Aldh1L1 JD130, in which the Aldh1L1 BAC drives expression of the eGFP-RpL10a (TRAP) construct for transcriptome profiling, we have previously shown that all Gfap+ cells express Aldh1L1 (Doyle et al. 2008). In the SVZ, this transgene is expressed in the same pattern as in Prism JD1849, as is the endogenous Aldh1L1 protein, demonstrating the expression pattern is not an artifact of transgenesis (Figure 2B). To systematically determine if the SVZ Aldh1L1 cells include neural stem cells, we next examined the expression of neural stem cell markers with a comprehensive transcriptomic approach.
Figure 2. Aldh1L1 is expressed in neurogenic regions in vivo.
A) In mouse line Prism JD1849, in which an Aldh1L1 BAC promoter drives expression of Cerulean, expression is seen in parenchemal astrocytes, as expected, but also in the neurogenic SVZ region (dotted lines) adjacent to lateral ventricles, and choroid plexus (**). B) In bacTRAP mouse line ALdh1L1 JD130, protein (red) labeling is consistent with Aldh1L1 BAC Egfp-L10a expression in all regions, including SVZa (dashed line). All scale bars = 50 micron
Comparative analysis of forebrain astrocytes reveals enrichment for neural stem cell and forebrain development genes
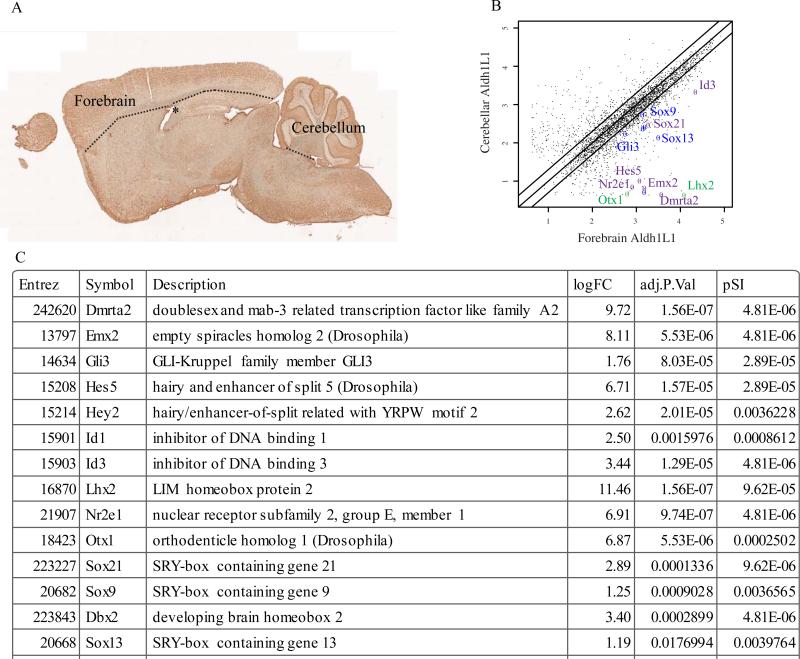
Previously, we utilized the TRAP to profile transcripts from Aldh1L1 expressing cells from multiple regions in the brain (Doyle et al. 2008): notably the cortex/forebrain and the cerebellum, with the former dissection including the dorsal surface of the lateral ventricles and some ventricular zone (Figure 3A). Comparative analysis of these two regions reveals 127 transcripts significantly enriched in Aldh1L1 expressing cells from the forebrain by at least 2 fold (Figure 3B) using conservative criteria. Careful examination of this list revealed a robust enrichment of several transcription factors, such as Nr2e1(Tlx1), Sox21, Id1, Hes5, Dmrta2, Emx2, and Lhx2 which are known to regulate either telencephalic development and/or adult neurogenesis (Boncinelli et al. 1993; Galli et al. 2002; Hatakeyama et al. 2004; Lyden et al. 1999; Monuki et al. 2001; Qu et al. 2010; Sandberg et al. 2005; Yoshizawa et al. 2011) (Figure 3C). Two of these(Nr2e1 and Lhx2) were also previously shown to be enriched in astrocytes cultured specifically from the forebrain(Yeh et al. 2009). To determine if this enrichment in neural stem cell genes was statistically significant, we compared the current analysis to our former screen for neural stem cell genes (Geschwind et al. 2001). The ~50 genes discovered in that early screen were significantly overlapped with the 127 genes discovered here(p<.0001, Fisher Exact Test) and included the functionally validated Psph (Nakano et al. 2007).
Figure 3. Forebrain astrocytes express neural stem cell genes in vivo.
A) Sagittal section of adult mouse brain from TRAP line Aldh1L1 JD130 stained with anti-GFP antibody shows Aldh1L1 BAC activity throughout the brain. Forebrain/cortical dissection included the corpus callosum and ventricular zone of the lateral ventricles. B) Scatterplot of microarray data(log scale) shows robust enrichment of known factors involved in regulation of neurogenesis (purple) or forebrain development (green), as well as new candidate factors identified here (blue). Black lines are 0.5, 1, and 2 fold. Median of three experiments, showing only those genes scored as expressed (see methods). C) Table highlighting several of the transcription factors enriched at least two fold in forebrain astrocytes. Complete analyzed data are available in Supplemental Table 1. logFC: log base 2 of the ratio of forebrain to cerebellar astrocyte gene expression. Adj.P.Val: B-H corrected p-value comparing forebrain to cerebellar astrocytes. pSI: Specificity index statistic for the relative enrichment of the transcript in forebrain neurons compared to all other neural cell types collected.
In contrast, it is worth noting that we do not see enriched expression of a large set of proliferation genes such as the Cyclins, Pcna, or Ki-67 nor related markers of the transient amplifying cells such as Eomes(Tbr2) (Englund et al. 2005), Ascl1(Mash1) (Parras et al. 2004), or Pbk (Dougherty et al. 2005). Likewise, in our previous profiling of Pdgfra expressing ‘oligodendrocyte progenitor cells’ from a similar dissection, we did see robust enrichment of cyclins, and proliferating progenitor genes such as Pbk, but not of Aldh1L1, nor the neurogenesis markers described above (Dougherty et al. 2012a). This suggests that relatively quiescent Aldh1L1+ cells give rise to a more rapidly proliferating Aldh1L1− neural progenitor, which then differentiates into the neuroblasts of the rostral migratory stream. To confirm this, we utilized Cre-Lox mediated fate mapping to examine the fate of Aldh1L1 expressing cells.
Fate mapping indicates Aldh1L1 is expressed in postnatal neural stem cells in vivo
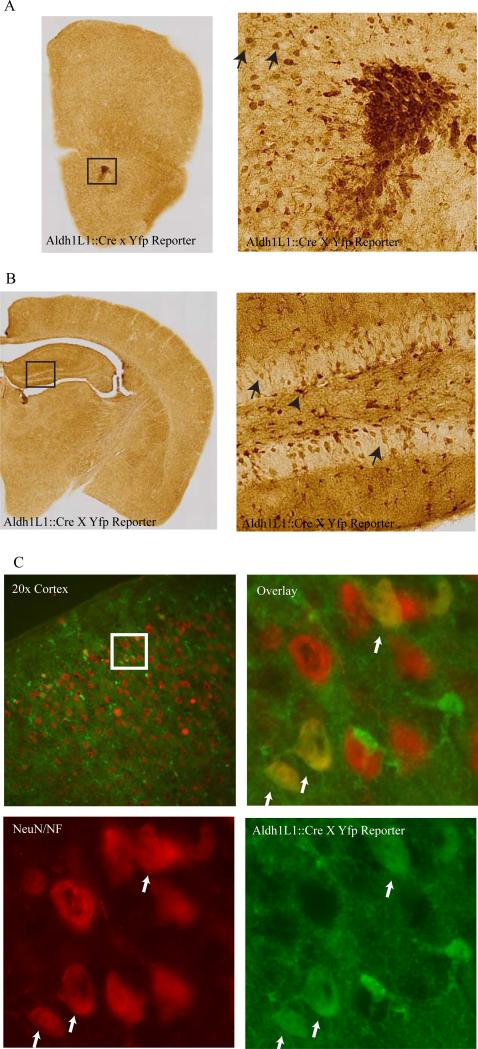
Using the same Aldh1L1 BAC, we generated multiple lines of mice expressing the Cre recombinase (Tien et al. 2012). Crossing these to a reporter line revealed a robust labeling of astrocytes throughout the brain, as expected, as well as clear labeling of cells in the rostral migratory stream with the morphology of neuroblasts (Figure 4A). Likewise in the hippocampus, there was robust labeling in SGZ astrocytes, and sporadic labeling of neurons in the granule cell layer of the dentate gyrus, consistent with Cre activity in hippocampal neural stem cells as well (Figure 4B).
Figure 4. Lineage tracing with Aldh1L1 Cre mice labels neurons.
A) Anti-GFP antibody staining of mouse forebrain from and Aldh1L1 Cre line JD1884, crossed to a YFP reporter mouse, shows expression in glia throughout the parenchema, and in presumptive neuroblasts of the rostral migratory stream (box, right panel) and adjacent neurons in the olfactory bulb. B) In the neurogenic dentate gyrus (box, right panel), GFP is seen in glial cells in the SGZ (arrowhead) and neurons of the granule cell layer (arrows). C) Immunofluorescence for YFP (green) and a mix of neuronal markers (NeuN and NF200, red), reveals some upper layer cortical neurons also derive from Aldh1L1 expressing cells (white arrows).
Outside of the neurogenic zones, we also saw sporadic labeling in cells with neuronal morphology (for example, in the upper layers of cortex). Double labeling with neuronal markers NeuN and NF200 confirms that Aldh1L1 expressing cells can give rise to some cortical neurons (Figure 4C), likely due to the reported expression of Aldh1L1 in some radial glia during development (Anthony and Heintz 2007). This same pattern of expression was reproduced to varying degrees across multiple lines generated by us, though we did note a surprising variability in neuronal expression even between littermates of the same line. Some neuronal labeling is evident as well in the Aldh1L1 CRE line generated by GENSAT(www.gensat.org). In contrast, we did not see substantial overlap of Aldh1L1 with the oligodendrocyte marker Mbp in the cortex suggesting Aldh1L1 expressing cells do not give rise to this cell type in this tissue. In the spinal cord, there was some small percentage of Mbp cells labeled in the Cre lines, but no neurons, in line JD1884 (Tien et al. 2012).
Discussion
The utility of Aldh1L1 as a marker for astrocytes
Given the knowledge that a subset of astrocytes in vivo serve as neural stem cells, it is perhaps unsurprising that Aldh1L1 is expressed in these cells as well. It is important to note that our studies here do not preclude the use of Aldh1L1 as a marker of astrocytes in the mature brain. With both our transgenic lines and antibodies we saw robust and apparently complete labeling of astrocytes throughout the CNS. In the non-Cre lines, in all regions we saw perfect concordance of Aldh1L1 BAC expression and Aldh1L1 protein, with the notable exception of some neurons in the anterior dorsal thalamus that were transgene positive, but antibody negative in multiple transgenic lines(not shown). However, genetic studies employing Aldh1L1 driven Cre for astrocyte specific lineage studies will have to contend with the caveats presented here – existing Aldh1L1 driven Cre lines will also recombine in post-natal neural stem cells and in some radial glial derived neurons, with variable penetrance. This does not preclude the use of these lines as astrocyte-specific in some situations – such as for Cre-responsive viral reagents delivered postnatally. It also suggests that the same BAC may be more effectively used in combination with inducible Cre constructs (such as Cre-ERT2) later in development to avoid labeling the early derived neuronal populations, though even these may lead to recombination in Aldh1L1 expressing cells in the periphery, such as in the liver or Schwann cells.
In vitro the expression of Aldh1L1 does not particularly map to the mature astrocyte phenotype, with robust transgene expression in EGF/bFGF-containing neural stem cell culture conditions, and relatively poor expression in ‘standard’ serum containing astrocyte media. It would be interesting to determine how the transgene responds in new HBEGF culture conditions optimized to culture the more mature astrocyte (Foo et al. 2011).
Gene expression in forebrain astrocytes
One surprising facet of our analysis here was the robust differences in astrocyte gene expression between the forebrain (containing neural stem cells) and the cerebellum. As there were also a large number of genes found enriched in astrocytes of the cerebellum, the differences seen cannot be accounted for entirely by the presence of the neurogenic niche in the forebrain. It is possible that these are region specific factors, or potentially factors needed for the specialized giant astrocyte-like cell of the cerebellum: the Bergmann glia. These cells also express Aldh1L1, but have a distinct morphology and gene expression from other astrocyte-like cells (Doyle et al. 2008; Zhang and Barres 2010).
In the forebrain data, our comparative analysis identified several factors known to be important in adult neurogenesis. This suggests that others genes from this analysis may be equally important for astrocytes in the neurogenic niche. For example, it is well known that Notch signaling is essential for maintenance of adult neurogenesis (Imayoshi and Kageyama 2011), and our data reflect a preponderance of Notch signaling in astrocytes of the forebrain. Yet, there is a puzzle to these data – deletion of the Rbpj, a key component of all intracellular Notch signaling cascades, totally ablates adult neural stem cell maintenance in the telencephalon (Imayoshi et al. 2010), yet triple knockouts of the downstream transcription factors Hes1;Hes3;Hes5 do not recapitulate this phenotype in the telencephalon (Hatakeyama et al. 2004). Examination of our data suggests that Hes7 may also be expressed in forebrain astrocytes, and could be compensating for this loss. To facilitate studies in this direction, we have provided a more comprehensive list of the output of this analysis as Supplemental Table 1. Other genes not before studies in this context, such as Dbx2 or Notch4 might be equally important for adult neurogenesis.
Supplementary Material
Acknowledgements
The mouse lines utilized here were generated in the laboratory of Nathaniel Heintz with funding from the Adelson Medical Research Foundation and HHMI. We thank Juliet Zhang for technical assistance, David Rowitch, Hui-Hsin Tsai and Ben Barres for their assistance in the evaluation of Cre lines, Sergey Krupenko for Aldh1L1 antibody, and Kelly Monk for advice. This work was supported by the NIH(1R21NS083052-01), and the Mallinckrodt foundation.
References
- Alvarez-Buylla A, Garcia-Verdugo JM. Neurogenesis in adult subventricular zone. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:629–34. doi: 10.1523/JNEUROSCI.22-03-00629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony TE, Heintz N. The folate metabolic enzyme ALDH1L1 is restricted to the midline of the early CNS, suggesting a role in human neural tube defects. Journal of Comparative Neurology. 2007;500:368–383. doi: 10.1002/cne.21179. [DOI] [PubMed] [Google Scholar]
- Boncinelli E, Gulisano M, Broccoli V. Emx and Otx homeobox genes in the developing mouse brain. Journal of neurobiology. 1993;24:1356–66. doi: 10.1002/neu.480241008. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu FC, Goldman JE. Regulation of Glial Fibrillary Acidic Protein (Gfap) Expression in Cns Development and in Pathological States. Journal of Neuroimmunology. 1985;8:283–292. doi: 10.1016/s0165-5728(85)80067-9. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CCA, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Dai MH, Wang PL, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic acids research. 2005;33 doi: 10.1093/nar/gni179. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal J, Roh JH, Maloney SE, Akuffo A, Shah S, Yuan H, Akuffo A, Wamsley B, Jones WB, Strong CdG. Translational profiling of hypocretin neurons identifies Lhx9 as necessary for normal development of the hypocretinergic system. Genes & Development. doi: 10.1101/gad.207654.112. others. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F. The glial identity of neural stem cells. Nature neuroscience. 2003;6:1127–34. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- Dougherty JD, Fomchenko EI, Akuffo AA, Schmidt E, Helmy KY, Bazzoli E, Brennan CW, Holland EC, Milosevic A. Candidate Pathways for Promoting Differentiation or Quiescence of Oligodendrocyte Progenitor-like Cells in Glioma. Cancer Res. 2012a;72:4856–68. doi: 10.1158/0008-5472.CAN-11-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Garcia AD, Nakano I, Livingstone M, Norris B, Polakiewicz R, Wexler EM, Sofroniew MV, Kornblum HI, Geschwind DH. PBK/TOPK, a proliferating neural progenitor-specific mitogen-activated protein kinase kinase. The Journal of neuroscience. 2005;25:10773–85. doi: 10.1523/JNEUROSCI.3207-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Maloney SE, Wozniak DF, Rieger MA, Sonnenblick L, Coppola G, Mahieu NG, Zhang J, Cai J, Patti GJ. The Disruption of Celf6, a Gene Identified by Translational Profiling of Serotonergic Neurons, Results in Autism-Related Behaviors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:2732–2753. doi: 10.1523/JNEUROSCI.4762-12.2013. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic acids research. 2010;38:4218–30. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Zhang J, Feng H, Gong S, Heintz N. Mouse transgenesis in a single locus with independent regulation for multiple fluorophores. PLoS ONE. 2012b;7:e40511. doi: 10.1371/journal.pone.0040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–62. doi: 10.1016/j.cell.2008.10.029. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP-thirty-one years (1969-2000). Neurochemical Research. 2000;25:1439–1451. doi: 10.1023/a:1007677003387. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:247–51. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu C, Barres BA. Regulation of synaptic connectivity by glia. Nature. 2010;468:223–31. doi: 10.1038/nature09612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC, Allen NJ, Bushong EA, Ventura PB, Chung WS, Zhou L, Cahoy JD, Daneman R, Zong H, Ellisman MH. Development of a method for the purification and culture of rodent astrocytes. Neuron. 2011;71:799–811. doi: 10.1016/j.neuron.2011.07.022. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R, Fiocco R, De Filippis L, Muzio L, Gritti A, Mercurio S, Broccoli V, Pellegrini M, Mallamaci A, Vescovi AL. Emx2 regulates the proliferation of stem cells of the adult mammalian central nervous system. Development. 2002;129:1633–44. doi: 10.1242/dev.129.7.1633. [DOI] [PubMed] [Google Scholar]
- Garcia AD, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nature neuroscience. 2004;7:1233–41. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Ou J, Easterday MC, Dougherty JD, Jackson RL, Chen Z, Antoine H, Terskikh A, Weissman IL, Nelson SF. A genetic analysis of neural progenitor differentiation. Neuron. 2001;29:325–39. doi: 10.1016/s0896-6273(01)00209-4. others. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–50. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–48. doi: 10.1016/j.cell.2008.10.028. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Kageyama R. The role of Notch signaling in adult neurogenesis. Molecular neurobiology. 2011;44:7–12. doi: 10.1007/s12035-011-8186-0. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:3489–98. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G. Why new neurons? Possible functions for adult hippocampal neurogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:635–8. doi: 10.1523/JNEUROSCI.22-03-00635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupenko SA, Oleinik NV. 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth & Differentiation. 2002;13:227–236. [PubMed] [Google Scholar]
- Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O'Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–7. doi: 10.1038/44334. others. [DOI] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. The Journal of cell biology. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17528–32. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, Walsh CA. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron. 2001;32:591–604. doi: 10.1016/s0896-6273(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Nakano I, Dougherty JD, Kim K, Klement I, Geschwind DH, Kornblum HI. Phosphoserine phosphatase is expressed in the neural stem cell niche and regulates neural stem and progenitor cell proliferation. Stem Cells. 2007;25:1975–84. doi: 10.1634/stemcells.2007-0046. [DOI] [PubMed] [Google Scholar]
- Nakano I, Paucar AA, Bajpai R, Dougherty JD, Zewail A, Kelly TK, Kim KJ, Ou J, Groszer M, Imura T. Maternal embryonic leucine zipper kinase (MELK) regulates multipotent neural progenitor proliferation. The Journal of cell biology. 2005;170:413–27. doi: 10.1083/jcb.200412115. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neymeyer V, Tephly TR, Miller MW. Folate and 10-formyltetrahydrofolate dehydrogenase (FDH) expression in the central nervous system of the mature rat. Brain research. 1997;766:195–204. doi: 10.1016/s0006-8993(97)00528-3. [DOI] [PubMed] [Google Scholar]
- Parras CM, Galli R, Britz O, Soares S, Galichet C, Battiste J, Johnson JE, Nakafuku M, Vescovi A, Guillemot F. Mash1 specifies neurons and oligodendrocytes in the postnatal brain. The EMBO journal. 2004;23:4495–505. doi: 10.1038/sj.emboj.7600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrieger FW, Slezak M. Genetic approaches to study glial cells in the rodent brain. Glia. 2012;60:681–701. doi: 10.1002/glia.22283. [DOI] [PubMed] [Google Scholar]
- Qu Q, Sun G, Li W, Yang S, Ye P, Zhao C, Yu RT, Gage FH, Evans RM, Shi Y. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol. 2010;12:31–40. doi: 10.1038/ncb2001. sup pp 1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–10. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Rhodes KJ, Trimmer JS. Antibodies as valuable neuroscience research tools versus reagents of mass distraction. Journal of Neuroscience. 2006;26:8017–8020. doi: 10.1523/JNEUROSCI.2728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg M, Kallstrom M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nature neuroscience. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Sun W, McConnell E, Pare JF, Xu QW, Chen M, Peng WG, Lovatt D, Han XN, Smith Y, Nedergaard M. Glutamate-Dependent Neuroglial Calcium Signaling Differs Between Young and Adult Brain. Science. 2013;339:197–200. doi: 10.1126/science.1226740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien AC, Tsai HH, Molofsky AV, McMahon M, Foo LC, Kaul A, Dougherty JD, Heintz N, Gutmann DH, Barres BA. Regulated temporal-spatial astrocyte precursor cell proliferation involves BRAF signalling in mammalian spinal cord. Development. 2012;139:2477–87. doi: 10.1242/dev.077214. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. Control of synapse number by glia. Science. 2001;291:657–61. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- Yang Y, Vidensky S, Jin L, Jie C, Lorenzini I, Frankl M, Rothstein JD. Molecular comparison of GLT1+ and ALDH1L1+ astrocytes in vivo in astroglial reporter mice. Glia. 2011;59:200–7. doi: 10.1002/glia.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T, Lee da Y, Gianino S, Gutmann D. Microarray analyses reveal regional astrocyte heterogeneity with implications for neurofibromatosis type 1 (NF1)-regulated glial proliferation. Glia. 2009;57:1239–1249. doi: 10.1002/glia.20845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa A, Nakahara Y, Izawa T, Ishitani T, Tsutsumi M, Kuroiwa A, Itoh M, Kikuchi Y. Zebrafish Dmrta2 regulates neurogenesis in the telencephalon. Genes to cells : devoted to molecular & cellular mechanisms. 2011;16:1097–109. doi: 10.1111/j.1365-2443.2011.01555.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Barres BA. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol. 2010;20:588–94. doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.