Abstract
Tumor necrosis factor-alpha (TNF-α) is an important cytokine in generating an immune response against infection with hepatitis C virus (HCV). The functions of TNF-α may be altered by single-nucleotide polymorphisms (SNPs) in its gene structure. We hypothesized that SNPs in TNF-α may be important in determining the outcome of an HCV infection. To test this hypothesis, we investigated the role of the polymorphism -308G/A, which is located in the promoter region of the TNF-α gene, in the progression of HCV infection in Egyptian patients using a quantitative real-time polymerase chain reaction (qRT-PCR). The distribution of this polymorphism and its impact on the serum level of TNF-α was compared between 90 HCV-infected patients [45 with HCV-induced cirrhosis and 45 with HCV-related hepatocellular carcinoma (HCC)] and 45 healthy Egyptian volunteers without any history of liver disease. Our results showed that at the TNF-α -308 position, the G/G allele was most common (78.5%) in the study population, with the G/A and A/A alleles occurring less frequently (13.3% and 8.1%, respectively). Frequencies of G/G, G/A, and A/A genotypes were 87%, 7%, and 6% in patients with liver cirrhosis and were 94%, 4%, and 2% in patients with HCC, respectively. Serum levels of TNF-α were significantly higher in HCV-infected patients than in healthy controls, indicating that the TNF-α -308 polymorphism does not influence the production of TNF-α. The serum level of TNF-α was positively correlated with HCV infection. Taken together, these findings suggest that the TNF-α -308 polymorphism may not be a host genetic factor associated with the severity of HCV infection, but may be an independent risk factor for HCC.
Keywords: Tumor necrosis factor-α, polymorphism, hepatitis C virus, liver neoplasm
Hepatitis C virus (HCV), a hepatotropic, non-cytopathic, positive-strand RNA virus that belongs to the Flaviviridae family, is a major cause of chronic liver disease[1]. An estimated 180 million people are carriers of HCV[2]. Approximately 80% of infected patients fail to clear the virus and progress to chronic hepatitis. Some of those with chronic HCV infection may progress to liver cirrhosis and eventually hepatocellular carcinoma (HCC)[3], which ranks as the fifth most common cancer in the world[4]. Genotype 4 (HCV-G4) is the prevalent genotype in Egypt[5], the Kingdom of Saudi Arabia[6], and Africa[7]. Egypt has the highest HCV prevalence in the world, with an overall prevalence of 12% among the general population, 40% in persons above 40 years of age, and even higher among persons in rural areas[8]. Some studies have indicated a link between HCV-G4 and the frequency and development of HCC[9].
Cytokines, as the products of host response to inflammation, play an important role in the defense against viral infections[10]. The maximum capacity of cytokine production in individuals has a major genetic component[11]. Polymorphisms within the regulatory regions or signal sequences of cytokine genes have been shown to affect the overall expression and secretion of cytokines both in vitro and sporadically in vivo systems[12]. Associations between polymorphisms in cytokine genes and inflammation, allograft rejection, autoimmune, and infectious diseases have been reported[12]–[15].
Tumor necrosis factor-alpha (TNF-α), a potent antiviral cytokine with a wide range of proinflammatory activities[16], plays a pivotal role in host immune response to HCV infection because cytotoxic T lymphocytes (CTLs) in the liver secrete TNF-α[17]. Circulating TNF-α level increases during HCV infection[18]–[20], and hepatitis viral infection induces TNF-α production in human hepatocytes[21]. An elevated TNF-α level correlates with the severity of hepatic inflammation, fibrosis, and tissue injury[19],[22]–[24]. Persistent immune-mediated hepatic injury can initiate the process of fibrosis, cirrhosis, and, eventually, HCC[22],[25]–[27].
TNF-α expression is tightly controlled at the transcriptional and post-transcriptional levels[12]. Six diallelic polymorphisms in the TNF-α promoter, which are thought to affect TNF-α production, have been reported and occur at positions -1031, -863, -857, -376, -308, and -238[12],[28],[29]. Polymorphism in the human TNF-α promoter at -308[30] involves the substitution of adenosine for guanine in the uncommon alleles. A variety of infectious diseases and inflammatory disorders are associated with TNF-α -308 alleles[31]. The role of polymorphisms in TNF-α in the pathogenesis of HCV infection has been investigated, but some results are contradictory[32]–[36]. These discrepancies may be due to ethnic differences in the examined populations, leading to a differential distribution of cytokine gene polymorphisms.
We hypothesized that genetic variability in TNF-α -308 alleles may account for different susceptibility to HCV infection. Therefore, this study was designed to investigate the role of the -308G/A polymorphism in the TNF-α promoter in the progression of HCV infection in Egyptian patients.
Materials and Methods
Study population
Of consecutive Egyptian patients with chronic HCV infection who were admitted to the Tropical Medicine Department, Ain Shams University Hospital, Cairo, Egypt, 90 patients were studied: 45 had HCV-induced cirrhosis and 45 had HCC. As controls, 45 age-matched healthy Egyptian volunteers, with no history of liver disease, normal liver function tests, and no evidence of HCV infection (confirmed by PCR), were enrolled. Informed consent was obtained from all study subjects. The local ethics committee at Ain Shams University approved the study protocol. All investigations were performed in accordance with the Health and Human Ethical Clearance Committee guidelines for clinical studies, Menofiya University.
All participants were subjected to thorough history taking and clinical examinations. HCV antibody assay was performed using third-generation enzyme-linked immunosorbent assay (ELISA) (Murex Biotech Ltd.) and confirmed by reverse transcription-polymerase chain reaction using the Amplicore HCV assay (Roche Diagnostics Corp., Indianapolis, IN) and hepatitis B surface antigen (HBsAg) (Sorin Biomedica Co., Italy). All patient groups were HCV antibody- and HCV RNA-positive and were negative for HBsAg and schistosome infection. No patients underwent antiviral therapy, and none had a history of habitual alcohol consumption. Exclusion criteria included co-infection with hepatitis B virus (HBV) and/or schistosome infection.
Serologic testing
Serum levels of albumin, total bilirubin, direct bilirubin, alanine aminotransferase (ALT), alkaline phosphatase (ALP), and aspartate aminotransferase (AST) were tested. The diagnosis of cirrhosis was based on the presence of clinical manifestations of portal hypertension such as varices, encephalopathy, or ascites; biochemical abnormalities including elevated serum bilirubin, decreased serum albumin, or prolonged prothrombin time; and obvious morphologic change of the liver detected by hepatic imaging such as ultrasonography.
HCC diagnosis
The diagnosis of HCC was made after reviewing images generated with several imaging modalities. Nodules larger than 1 cm found in the ultrasound screening of a cirrhotic liver were investigated further with either 4-phase multidetector computed tomography (CT) scan or dynamic contrast-enhanced magnetic resonance imaging (MRI). If the appearances were typical of HCC, namely, hypervascular in the arterial phase with washout in the portal venous or delayed phase, the lesion was diagnosed as HCC. If the findings were not characteristic or the vascular profile was not typical, a second contrast-enhanced study with the other imaging modality was performed or the lesion was biopsied. All patients were confirmed not to have other cancers in an initial screening examination[37],[38].
DNA extraction
Genomic DNA was isolated from 200 µL of whole blood using the silica membrane spin column based kit (Qiagen, Hilden, Germany) according to manufacturer's instructions.
Polymorphism genotyping
TNF-α (-308G/A) SNP genotyping was performed using Assays-by-Design SM for SNP Genotyping (TaqMan® MGB probes, FAM™ and VIC® dye-labeled) (Applied Biosystems). The TNF-α -308 Custom TaqMan® SNP Genotyping assay (number 4331349) was performed in a 48-well plate format on a one-step unit (Applied Biosystems). Amplification was performed in a 25 µL volume reaction. The following cycling conditions were used: 10 min at 94°C, 15 s at 92°C, and 1 min at 60°C for 40 cycles.
TNF-α measurement
Serum levels of TNF-α were quantified using an ELISA that follows the quantitative sandwich immunoassay technique. This assay used immobilized monoclonal antibody and biotin-linked polyclonal antibody, both of which were specific for human TNF-α. Commercially available matched and paired antibodies were used (R&D Systems Inc. Minneapolis, MN, USA). A total of 50 µL of anti-human monoclonal antibody (4 g/mL) was coated onto each well of a 96-well flat bottom microtiter plate (Griener Labortechnik, Kremsmunster, Austria) and incubated for 1 h at 37°C, then overnight at 4°C in a humidified chamber. Plates were washed three times with washing buffer, phosphate-buffered saline (PBS)/0.05% polyoxyethylene-20 (Tween-20), and blocked with 200 µL/well blocking buffer, PBS/0.05% Tween-20/5% fetal bovine serum (FBS) (Sigma Co., St. Louis, MO, USA) at 37°C for 1.5 h. For TNF-α level quantification, triplicate assays of 50 µL aliquots of patient serum samples and recombinant human TNF-α standards (R&D Systems) were incubated for 1 h at 37°C. At the end of the incubation, the plates were washed three times with washing buffer and incubated in diluted secondary antibody, biotin-labeled anti-human TNF-α polyclonal antibody (200 ng/mL), for 1 h at 37°C. After washing away any unbound substances, 50 µL of peroxidase-conjugated streptavidin (Jackson Immun-search Lab, USA), which was diluted at 1:1000, was added to each well, and the plates were incubated for 1 h at 37°C. After intensive washing, the enzyme reaction was carried out by adding 50 µL of substrate solution [equal volumes of 3,3′,5,5′-tetramethyl benzidine (TMB; 0.4 g/L) and H2O2 (0.02% in citric acid buffer; KPL, Kirkegaard and Perry Lab, Gaithersburg, MD, USA) to each well. Color development was stopped by addition of 50 µL/well of stopping buffer (1 mol/L HCI) (Surechern Products, Needham Marker, Suffolk, England). The intensity of the developed color was measured by reading optical absorbance at 450 nm using a microplate reader (FLUOstar OPTIMA, BMG LABTECH GmbH, Offenburg, Germany). The microplate reader software (Softmax) readily processed the raw absorbance values into a standard curve from which TNF-α concentration of unknown samples could be derived directly.
Statistical analysis
All statistical analyses was performed using the Statistical Package for Social Science (SPSS) version 10 (LEAD Technology Inc). Data are presented as means with corresponding standard error (SE). Comparisons among different groups were performed by one-way analysis of variance (ANOVA). The Tukey test was used as a post-hoc test. Frequency was compared between groups using the Chi-square test. Correlation between variables was determined using Pearson's correlation test. In all tests, the level of significance was P < 0.05.
Results
Demographic and clinical information of patients and controls
The demographic characteristics of the subjects enrolled are given in Table 1. Of the 90 HCV-infected patients, 69 were men and 21 were women, with an age ranged from 42 to 70 years; 45 had cirrhosis, and 45 had HCC. Of the 45 healthy controls, 21 were men and 24 were women, with an age ranged from 34 to 70 years, without any history of liver disease. The age of the patients was positively correlated to the progression of the disease (r=0.526, P < 0.001).
Table 1. Demographic and clinical characteristics of the studied population.
| Parameter | Healthy controls (n = 45) | Cirrhosis patients (n = 45) | HCC patients (n = 45) | P | Correlation with disease progression |
| Gender (♂/♀) | 21/24 | 33/12 | 36/9 | <0.01 | |
| Age (years) | 44 ± 3 | 54 ± 2 | 58 ± 2 | <0.001 | r= 0.526, P<0.001 |
| (34-70) | (42-62) | (43-70) | r= 0.480, P<0.001 | ||
| Comma(pre / negative) | Negative | 15/30 | 24/21 | <0.001 | |
| Ascites (moderate/negative) | Negative | 18/27 | 18/27 | <0.001 | r= 0.369, P<0.001 |
| INR | 1.16 ±0.03 | 1.25 ±0.03 | 1.48 ± 0.05 | <0.001 | r= 0.443, P<0.001 |
| ALT (IU/L) | 21.66 ±0.76 | 43.53 ± 2.77 | 67.33 ± 4.88 | <0.001 | r= 0.650, P<0.001 |
| AST(IU/L) | 28.66 ± 1.04 | 68.95 ± 4.48 | 90.08 ± 5.54 | <0.001 | r= 0.679, P<0.001 |
| ALP (IU/L) | 68.46 ±1.59 | 86.66 ± 3.40 | 99.53 ± 4.57 | <0.001 | r= 0.488, P<0.001 |
| Albumin (g/L) | 4.13 ± 0.08 | 2.66 ± 0.11 | 2.78 ± 0.10 | <0.001 | r= -0.615, P<0.001 |
| Bilirubin | |||||
| Total (µmol/L) | 0.94 ± 0.04 | 1.70 ±0.10 | 1.64 ± 0.10 | <0.001 | r= 0.423, P<0.01 |
| Direct (µmol/L) | 0.16 ± 0.01 | 0.75 ± 0.06 | 0.50 ± 0.04 | <0.001 | r= 0.382, P<0.01 |
| HCV | |||||
| Ab | Negative | Positive | Positive | ||
| PCR | Negative | Positive | Positive | ||
| HBsAg | Negative | Negative | Negative |
HCC, hepatocellular carcinoma; ALT, alinine aminotransferase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; INR, international normalization ratio; HCV, hepatitis C virus. All data are presented as mean ± standard error (SE). Differences among the 3 groups were compared by ANOVA test.
TNF-α genotype
The distribution of genotypes for TNF-α at position -308 are given in Table 2. The SNP frequency at the TNF-α -308 position was significantly different between HCV-infected patients and healthy individuals (P<0.001). On the other hand, no significant difference in the SNP frequency at the TNF-α -308 position was recorded between patients with cirrhosis or HCC. The frequency of the G allele was significantly higher in HCV-infected patients than in healthy controls (P<0.05), whereas the frequency of the A allele was significantly higher in healthy controls than in HCV-infected patients (P<0.001).
Table 2. Distribution of TNF-α genotypes and frequencies in HCV-infected patients and healthy controls.
| Genotype | Healthy controls (n = 45) | Cirrhosis patients (n = 45) | HCC patients (n = 45) |
| -308 G/A genotype | |||
| G/G | 25 (56) | 39 (87)a | 42 (94)a |
| G/A | 13 (29) | 3 (7)b | 2 (4)a |
| A/A | 7 (15) | 3 (6) | 1 (2)c |
| GA+AA | 38 (84) | 42 (93)a | 44 (97)a |
| -308 G/A alleles | |||
| G | 63 (70) | 81 (90) | 86 (96)c |
| A | 27 (30) | 9 (10)a | 4 (4)a |
All data are presented as cases (%). aP< 0.001, bP < 0.01, cP < 0.05, vs. controls.
HCV infection and serum TNF-α level
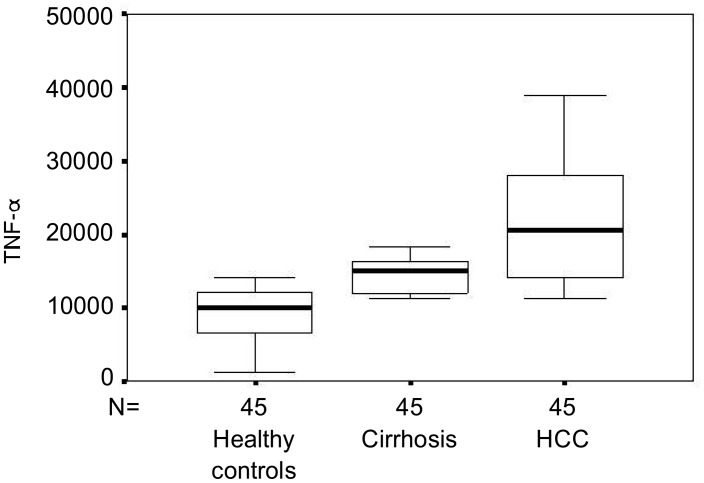
Compared with those in healthy controls, serum TNF-α levels were gradually elevated in HCV-infected patients (P<0.001) (Figure 1). Patients with HCC had significantly higher TNF-α levels than patients with cirrhosis (P<0.001). The serum level of TNF-α was positively correlated to the severity of the HCV infection (r=0.691, P<0.001).
Figure 1. Serum levels of tumor necrosis factor-α (TNF-α) in hepatitis C virus (HCV)-infected patients. Data are presented as box plots with lines inside the box representing medians, boxes representing 25th and 75th percentiles and the lines outside the boxes indicating 10th and 90th percentiles. HCC, hepatocellular carcinoma.
Discussion
The promoter of TNF-α has many biallelic variations, and the one at position -308 (G-308A), relative to the transcription start site[39], has been linked with several inflammatory, autoimmune, infectious, and malignant diseases[40]. The impact of host genetic factors and the polymorphism at the -308 position on the clinical outcome of HCV infection have not been fully elucidated. Thus, this study examined the influence of genetic variation on the regulation of TNF-α and disease progression.
The role of polymorphisms in proinflammatory cytokines is unclear. Although one report indicates that the TNF2 allele is more frequently found in patients with cirrhosis than in those with less severe liver disease[32], another study has not confirmed these results[41]. In the current study, we found significant difference in the distribution of TNF-α promoter polymorphism at position -308 between HCV-infected patients enrolled in this work. Our results are consistent with those reported by Zein et al.[42], but quite different from another study[43]. SNPs at -308 showed inconsistent associations with various HCV outcomes[32],[44],[45]. The -308A promoter variant has been associated with enhanced transcription of TNF-α [12], but other investigations have failed to confirm this finding[46]. In a previous study, no significant difference was observed in the frequency of the TNF-α -308.2 allele at position -308, which had been linked formerly to higher TNF-α production[12]. Likewise, another study did not find the SNP at -308 to be associated with viral recovery or persistence[35]. Yee et al.[32] found that -308A was associated with increased cirrhosis in HCV-infected patients.
In considering the relationship between TNF-α -308 polymorphisms and the severity of disease, Barrett et al.[35] and Hohler et al.[45] reported an association between these factors in patients with chronic HCV infection; Romero-Gomez et al.[47] found no association between TNF-α -308 polymorphisms and the severity of fibrosis in HCC. Hence, studies of TNF-α -308 polymorphisms and its effect on TNF-α production have been inconsistent but nevertheless indicate that this position likely influences the production of TNF-α, which may in turn affect the outcome of an HCV infection. Similarly, functional studies of the allele -308A polymorphism have been inconsistent, with both higher[48] and unchanged[49] TNF-α production in cells stimulated with lipopolysac-charide. Yee et al. [32] showed that TNF-α -308.2 conferred a 5.1-fold risk of cirrhosis for patients with chronic HCV infection. No correlation between TNF-α -308 promoter polymorphisms and necroinflammatory histological activity was previously demonstrated.
Hepatocyte damage elicits an inflammatory response through activation of tissue macrophage Kupffer cells. These activated cells release an array of antiviral cytokines including TNF-α, which plays a pivotal role in host immune response to HCV infection. In this study, elevated TNF-α levels were seen in HCV-infected patients with HCC compared to healthy controls. We previously reported elevation of TNF-α levels in HCV-infected patients[50]. These results are consistent with known mechanisms involved in the progression of chronic HCV infection, including activation of proinflammatory cytokines by the virus[11]. In accordance with our results, circulating TNF-α level has been reported to increase during HCV infection[18]–[20],[22]. An elevated TNF-α level has been correlated with the severity of hepatic inflammation, fibrosis, and tissue injury[19],[22]–[24]. Moreover, TNF-α plays an important role in hepatic fibrogenesis and progression of fibrosis in chronic liver disease[22],[24].
Our results showed a significant elevation in serum TNF-α levels in patients with HCC, well above those of patients with cirrhosis. Chronic HCV infection has been well established as an independent risk factor for HCC[34],[35]. As with oncogenic viruses, chronic HCV infection may lead to persistent hepatocyte necroinflammation and hepatic fibrosis[17],[26]–[27]. Persistent immune-mediated hepatic injury can initiate the process of fibrosis, cirrhosis, and, eventually, HCC[17],[22],[25]–[27]. In conclusion, this preliminary study indicated that inheritance of the TNF-α promoter genotype at position -308 is not associated with clinical features of HCV infection. However, one of the shortcomings of this study is its rather small sample size; therefore, the results should be confirmed in a larger series, as well as in patients of different ethnic origins. Based on these findings, further analysis of polymorphisms in other immune response genes in a large study population is required to understand the consequence of gene variants in HCV infection.
References
- 1.Shepard C, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5(9):558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Dai C, Chuang W, Chang W, et al. Tumor necrosis factor alpha promoter polymorphism at position-308 predicts response to combination therapy in hepatitis C virus infection. J Infect Dis. 2006;193(1):98–101. doi: 10.1086/498244. [DOI] [PubMed] [Google Scholar]
- 3.Lauer G, Walker B. Hepatitis C virus infection. N Engl J Med. 2001;345(1):41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 4.Donato F, Gelatti U, Limina R, et al. Southern Europe as an example of interaction between various environmental factors: a systematic review of the epidemiologic evidence. Oncogene. 2006;25(27):3756–3770. doi: 10.1038/sj.onc.1209557. [DOI] [PubMed] [Google Scholar]
- 5.Egyptian Ministry of Health Annual report. 2007 Available at http://www.mohp.gov.eg/Main.asp. [Google Scholar]
- 6.Shobokshi O, Serebour F, Skakni L, et al. Hepatitis C genotypes and subtypes in Saudi Arabia. J Med Virol. 1999;58(1):44–48. [PubMed] [Google Scholar]
- 7.Simmonds P. Genetic diversity and evolution of hepatitis C virus—15 years on. J Gen Virol. 2004;85(Pt 11):3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 8.Nafeh M, Abdel-Hamid M, Strickland G, et al. Hepatitis C in a community in Upper Egypt: risk factors for infection. Am J Trop Med Hyg. 2002;66(5):633–638. doi: 10.4269/ajtmh.2002.66.633. [DOI] [PubMed] [Google Scholar]
- 9.Abdel Hamid M, El Daly M, Molnegren V, et al. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J Gen Virol. 2007;88(5):1526–1531. doi: 10.1099/vir.0.82626-0. [DOI] [PubMed] [Google Scholar]
- 10.Koziel M. Cytokines in viral hepatitis. Semin Liver Dis. 1999;19(2):157–169. doi: 10.1055/s-2007-1007107. [DOI] [PubMed] [Google Scholar]
- 11.Fan XG, Liu WE, Li CZ, et al. Circulating Th1 and Th2 cytokines in patients with hepatitis C virus infection. Mediators Inflamm. 1998;7(4):295–297. doi: 10.1080/09629359890992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson A, Symons J, McDowell T, et al. Effects of a polymorphismin the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94(7):3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Ari Z, Mor E, Papo O, et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98(1):144–150. doi: 10.1111/j.1572-0241.2003.07179.x. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Mestre M, Gendzekhadze K, Rivas-Vetencourt P, et al. TNF-alpha-308 A allele, a possible severity risk factor of hemorrhagic manifestation in dengue fever patients. Tissue Antigens. 2004;64(4):469–472. doi: 10.1111/j.1399-0039.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- 15.Bendicho M, Guedes J, Silva N, et al. Polymorphism of cytokine genes (TGF-beta1, IFN-gamma, IL-6, IL-10, and TNFalpha) in patients with chronic pancreatitis. Pancreas. 2005;30(4):333–336. doi: 10.1097/01.mpa.0000161809.24284.33. [DOI] [PubMed] [Google Scholar]
- 16.Beutler BA. The role of tumor necrosis factor in health and disease. J Rheumatol. 1999;26(Suppl 57):16–21. [PubMed] [Google Scholar]
- 17.Koziel M, Dudley D, Afdhal N, et al. HLA class l-restricted cytotoxic T lymphocytes specific for hepatitis C virus—identification of multiple epitopes and characterization of patterns of cytokine release. J Clin Invest. 1995;96(5):2311–2321. doi: 10.1172/JCI118287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odeh M, Sabo E, Srugo I, et al. Relationship between tumor necrosis factor-α and ammonia in patients with hepatic encephalopathy due to chronic liver failure. Ann Med. 2005;37(8):603–612. doi: 10.1080/07853890500317414. [DOI] [PubMed] [Google Scholar]
- 19.Falasca K, Ucciferri C, Dalessandro M, et al. Cytokine patterns correlate with liver damage in patients with chronic hepatitis B and C. Ann Clin Lab Sci. 2006;36(2):144–150. [PubMed] [Google Scholar]
- 20.Cua I, Hui J, Bandara P, et al. Insulin resistance and liver injury in hepatitis C is not associated with virus-specific changes in adipocytokines. Hepatology. 2007;46(1):66–73. doi: 10.1002/hep.21703. [DOI] [PubMed] [Google Scholar]
- 21.González-Amaro R, García-Monzón C, García-Buey L, et al. Induction of tumor necrosis factor a production by human hepatocytes in chronic viral hepatitis. J Exp Med. 1994;179(3):841–848. doi: 10.1084/jem.179.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang E, Del Vecchio A, Smolinski S, et al. Biomedicines to reduce inflammation but not viral load in chronic HCV: what's the sense? Trends Biotechnol. 2004;22(10):517–523. doi: 10.1016/j.tibtech.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Akpolat N, Yahsi S, Godekmerdan A, et al. Relationship between serum cytokine levels and histopathological changes of liver in patients with hepatitis B. World J Gastroenterol. 2005;11(21):3260–3263. doi: 10.3748/wjg.v11.i21.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamal S, Turner B, He Q, et al. Progression of fibrosis in hepatitis C with and without schistosomiasis: correlation with serum markers of fibrosis. Hepatology. 2006;43(4):771–779. doi: 10.1002/hep.21117. [DOI] [PubMed] [Google Scholar]
- 25.Thimme R, Wieland S, Steiger C, et al. CD8+ T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77(1):68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cha C, DeMatteo R. Molecular mechanisms in hepatocellular carcinoma development. Best Pract Res Clin Gastroenterol. 2005;19(1):25–37. doi: 10.1016/j.bpg.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene. 2006;25(27):3834–3847. doi: 10.1038/sj.onc.1209562. [DOI] [PubMed] [Google Scholar]
- 28.Bidwell J, Keen L, Gallagher G, et al. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun. 1999;1(1):3–19. doi: 10.1038/sj.gene.6363645. [DOI] [PubMed] [Google Scholar]
- 29.Negoro K, Kinouchi Y, Hiwatashi N, et al. Crohn's disease is associated with novel polymorphisms in the 5′-flanking region of the tumor necrosis factor gene. Gastroenterology. 1999;117(5):1062–1068. doi: 10.1016/s0016-5085(99)70390-2. [DOI] [PubMed] [Google Scholar]
- 30.Wilson A, di Giovine F, Blakemore A, et al. Single base polymorphism in the human tumour necrosis factor alpha (TNF alpha) gene detectable by Ncol restriction of PCR product. Hum Mol Gene. 1992;1(5):353. doi: 10.1093/hmg/1.5.353. [DOI] [PubMed] [Google Scholar]
- 31.Bidwell J, Keen L, Gallagher G, et al. Cytokine gene polymorphism in human disease: on-line databases, supplement 1. Genes Immun. 2001;2(2):61–70. doi: 10.1038/sj.gene.6363733. [DOI] [PubMed] [Google Scholar]
- 32.Yee L, Tang J, Herrera J, et al. Tumor necrosis factor gene polymorphisms in patients with cirrhosis from chronic hepatitis C virus infection. Genes Immun. 2000;1(6):386–390. doi: 10.1038/sj.gene.6363696. [DOI] [PubMed] [Google Scholar]
- 33.Tambur A, Ortegel J, Ben-Ari Z, et al. Role of cytokine gene polymorphism in hepatitis C recurrence and allograft rejection among liver transplant recipients. Transplantation. 2001;71(10):1475–1480. doi: 10.1097/00007890-200105270-00020. [DOI] [PubMed] [Google Scholar]
- 34.Vidigal P, Germer J, Zein N. Polymorphisms in the interleukin-10, tumor necrosis factor-alpha, and transforming growth factor-beta1 genes in chronic hepatitis C patients treated with interferon and ribavirin. J Hepatol. 2002;36(2):271–277. doi: 10.1016/s0168-8278(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 35.Barrett S, Collins M, Kenny C, et al. Polymorphisms in tumor necrosis factor-alpha, transforming growth factor beta, interleukin-10, interleukin-6, interferon-gamma, and outcome of hepatitis C virus infection. J Med Virol. 2003;71(2):212–218. doi: 10.1002/jmv.10472. [DOI] [PubMed] [Google Scholar]
- 36.Abbas Z, Moatter T, Hussainy A, et al. Effect of cytokine gene polymorphism on histological activity index, viral load and response to treatment in patients with chronic hepatitis C genotype 3. World J Gastroenterol. 2005;11(42):6656–6661. doi: 10.3748/wjg.v11.i42.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryder S. Guidelines for the diagnosis and treatment of hepatocellular carcinoma (HCC) in adults. Gut. 2003;52(Suppl. 3):iii1–8. doi: 10.1136/gut.52.suppl_3.iii1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruix J, Sherman M. Management of hepatocellular carcinoma: Practice Guidelines Committee, American Association for the Study of Liver Diseases. Hepatology. 2005;42(5):1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 39.Hajeer A, Hutchinson I. TNF-alpha gene polymorphism: clinical and biological implications. Microsc Res Tech. 2000;50(3):216–228. doi: 10.1002/1097-0029(20000801)50:3<216::AID-JEMT5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 40.Thursz M. Genetic susceptibility in chronic viral hepatitis. Antiviral Res. 2001;52(2):113–116. doi: 10.1016/s0166-3542(01)00175-9. [DOI] [PubMed] [Google Scholar]
- 41.Powell E, Edwards-Smith C, Hay J, et al. Host genetic factors influence disease progression in chronic hepatitis C. Hepatology. 2000;31(4):828–833. doi: 10.1053/he.2000.6253. [DOI] [PubMed] [Google Scholar]
- 42.Zein N, Germer J, El-Zayadi A, et al. Ethnic differences in polymorphisms of tumor necrosis factor-α, interleukin-10, and transforming growth factor-β1 genes in patients with chronic hepatitis C virus infection. Am J Trop Med Hyg. 2004;70(4):434–437. [PubMed] [Google Scholar]
- 43.Ho S, Wang Y, Chen H, et al. Increased risk of developing hepatocellular carcinoma associated with carriage of the TNF2 allele of the –308 tumor necrosis factor-promoter gene. Cancer Causes Control. 2004;15(7):657–663. doi: 10.1023/B:CACO.0000036173.99930.75. [DOI] [PubMed] [Google Scholar]
- 44.Constantini P, Wawrzynowicz-Syczewska R, Clare M, et al. Interleukin-1, interleukin-10 and tumour necrosis factor-alpha gene polymorphisms in hepatitis C virus infection: an investigation of the relationships with spontaneous viral clearance and response to alpha-interferon therapy. Liver. 2002;22(5):404–412. doi: 10.1034/j.1600-0676.2002.01553.x. [DOI] [PubMed] [Google Scholar]
- 45.Hohler T, Kruger A, Gerken G, et al. Tumor necrosis factor alpha promoter polymorphism at position -238 is associated with chronic active hepatitis C infection. J Med Virol. 1998;54(3):173–177. doi: 10.1002/(sici)1096-9071(199803)54:3<173::aid-jmv5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 46.Brinkman B, Zuijdgeest D, Kaijzel E, et al. Relevance of the tumor necrosis factor alpha (TNFα)-308 promoter polymorphism in TNF-α gene regulation. J Inflamm. 1996;46(1):32–41. [PubMed] [Google Scholar]
- 47.Romero-Gomez M, Montes-Cano M, Otero-Fernandez M, et al. SLC11A1 promoter gene polymorphisms and fibrosis progression in chronic hepatitis C. Gut. 2004;53(3):446–450. doi: 10.1136/gut.2003.028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Louis E, Franchimont D, Piron A, et al. Tumor necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin Exp Immunol. 1998;113(3):401–406. doi: 10.1046/j.1365-2249.1998.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huizinga T, Westendorp R, Bollen E, et al. TNF-alpha promoter polymorphisms, production and susceptibility to multiple sclerosis in different groups of patients. J Neuroimmunol. 1997;72(2):149–153. doi: 10.1016/s0165-5728(96)00182-8. [DOI] [PubMed] [Google Scholar]
- 50.Talaat R. Soluble angiogenesis factors in sera of Egyptian patients with hepatitis C virus infection: correlation with disease severity. Viral Immunol. 2010;23(2):151–157. doi: 10.1089/vim.2009.0089. [DOI] [PubMed] [Google Scholar]