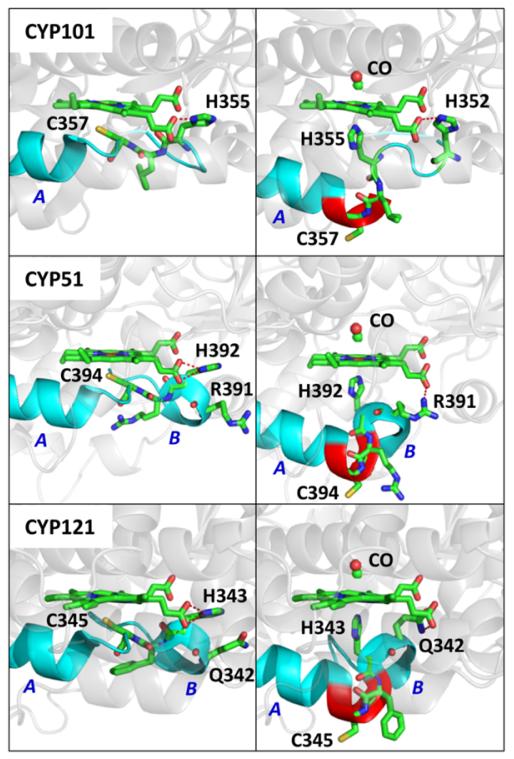
Figure 8.
The three panels on the left side show the crystal structure of CYP101 (PDB#2CPP), CYP51(PDB#1EA1), and CYP121(PDB#2IJ7). The L-helix is extended by 5 residues past the Cys ligand in the CYP51 and CYP121 structures. The helix structures on the left and right side of the proximal cysteine ligand are labeled as A and B, respectively. The red dashed line indicates the hydrogen bond between “near” propionate group and H355, H392 and H343 in the three structures, respectively. The three panels on the right show the possible CO bound P420 structures for the three proteins. The proposed structures are formed by extending the A helix by three amino acids (highlighted in red). A remnant of the B helix structure is preserved in the proposed P420-CO structure for CYP51 and CYP121. In the proposed structures, H352, R391, and Q342 replace the histidine residues of the native structures by forming a hydrogen bonds (red dashed line) with the propionate group. Larger renditions and different angles showing the possible structures underlying the ligand switch can be found in the SI.