Fig. 2.
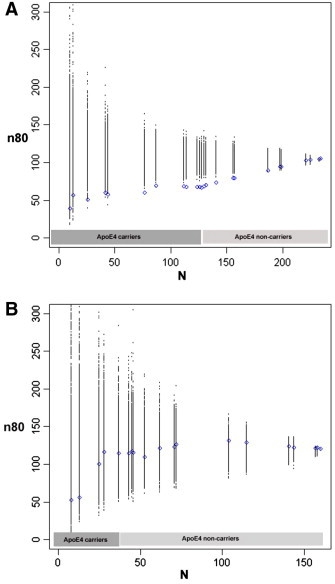
Sample size estimates (n80) are shown for hypothetical clinical trials as a function of number of subjects (N) after ranking subjects according to a cumulative genetic risk score derived from ApoE4 (coded as 0 or 1 for carrier status) and single nucleotide polymorphisms in CLU, CR1 and PICALM (blue). The breakpoints represent different risk score values, as these are not continuous (see Inline Supplementary Table S1 for a list of risk scores and corresponding allele combinations). Permutations are performed by randomizing the ranking procedure 1000 times and calculating n80 estimates for each N (all permuted estimates are shown in black). Panel A shows results for MCI subjects. Selection of subjects who carry at least one copy of the risk allele, ApoE4 (~ 55% of all MCI subjects), reduces n80s from 105 to 67. This is further reduced from the remaining three risk variants to 58 in the top ~ 20% and to 50 in the top ~ 10% of MCI subjects, respectively. Permutations consistently provide worse estimates. Panel B displays similar results for cognitively healthy subjects only. Selecting ApoE4 carriers (28% of all controls) reduces n80 minimally from 120 to 117. Selecting the top ~ 10% of control subjects with highest genetic risk reduces n80 to 55, still falling short of significance when compared to random permutations (p ~ 0.11).
