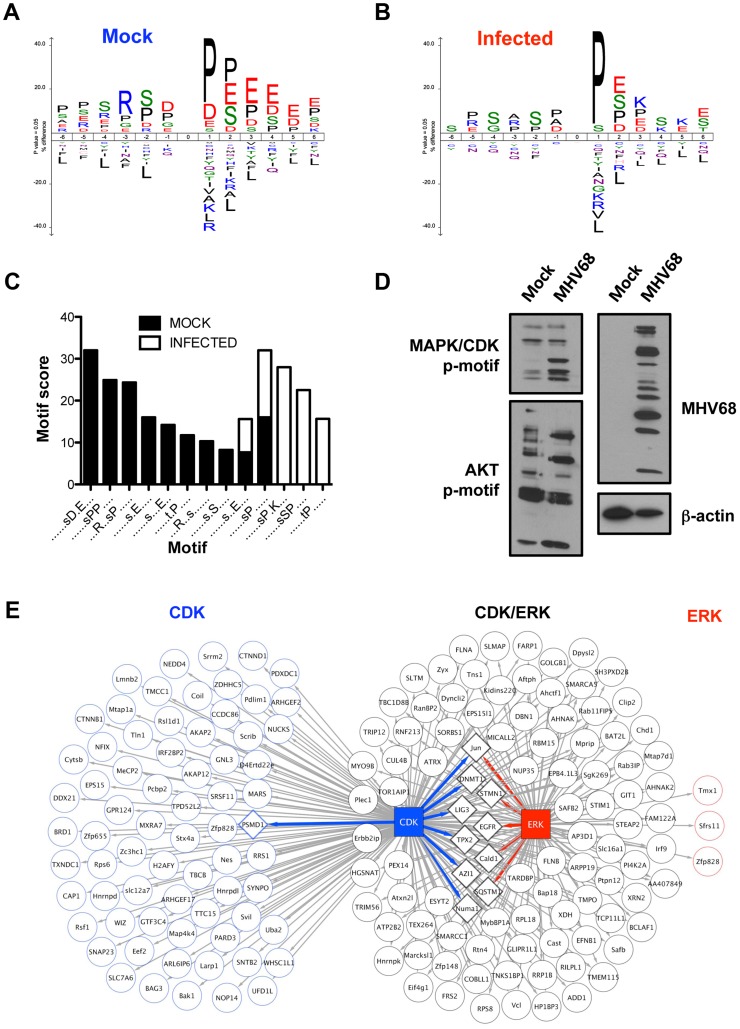
Figure 7. Comparative phosphorylation motif analyses reveal an infection-associated MAPK/CDK signature.
Unique phosphopeptides present in either mock-infected (A) or infected cells (B) were analyzed using ICE-LOGO. Weighted sequence context for residues flanking either p-Ser or p-Thr at position 0 is provided where a larger size designation corresponds to increased relative abundance of a particular amino acid in the global data set compared to other residues at the same position. (C) Unique phosphopeptides were analyzed using Motif-X to identify motifs over-represented relative to the Mus musculus background proteome at a p-value of <0.000001. (D) 3T3 fibroblasts were mock infected or infected with MHV68 at MOI = 5 PFU/cell. Cells were harvested 18 h post-infection, and proteins were resolved by SDS-PAGE. Immunoblot analyses were performed using antibodies directed against the indicated phosphorylation motifs or proteins. (E) Putative CDK1/2 and ERK1/2 phosphorylated proteins present in infected cells were identified by GPS 2.1 analysis on high confidence settings. A separate STRING analysis was performed to identify proteins that functionally interact with either CDK1/2 or ERK1/2. Arrows connect the kinase to its predicted substrate. STRING-defined substrate interactions are depicted as diamond-shaped nodes, where kinase-substrate interactions are color-coded blue or red to denote CDK or ERK connectivity, respectively.