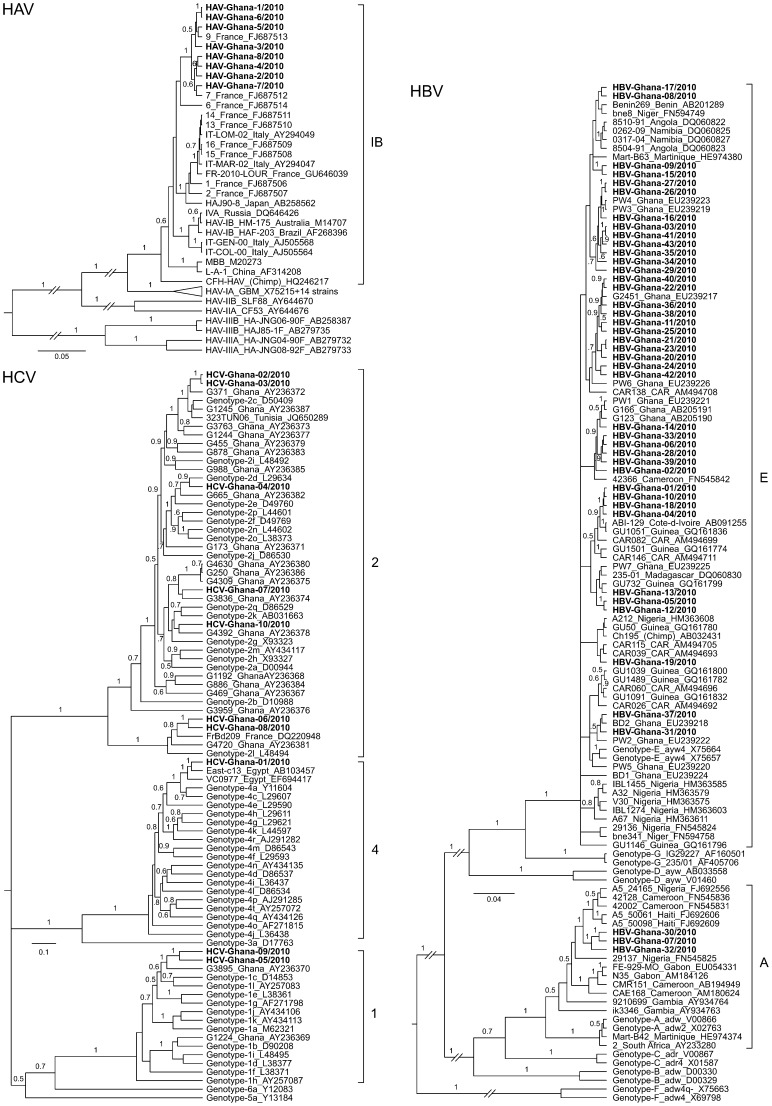
Figure 3. Phylogenetic analysis of HAV, HBV, and HCV sequences.
The alignments to reconstruct the phylogenies included the novel sequences of HAV (467 nucleotides of VP1/2A junction), HBV (684 nucleotides of N terminal P protein region), and HCV (337 nucleotides of NS5B region) as well as sequences available from GenBank by December 2012. The latter are identified by GenBank accession numbers. Sequences of this study are highlighted in boldface (GenBank accession no. KC632110–KC632153). Primers used for amplification and sequencing are shown in Table 1. Virus genotypes are indicated with the strains or right to the tree. If available, the geographic origin of a strain is indicated as well. Posterior probability values are shown on the branches if ≥0.5. Some branches with low support values were collapsed for clarity of presentation.