Table 1. Parameters of the computational model giving positive correlation with the experimental data on all simulated chromosomes.
| parameter | function | optimal range |
| pa | association rate at nucleation sites | 0.01–0.1 |
| pd | deletion rate | 0.01–0.02 |
| ps,1 | propagation rate of heterochromatin marks | ps,1 = 0.1–0.16 |
| ps,2 | propagation rate of euchromatin marks | ps,2 = 0.1 |
The propagation rate 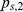 was left unchanged.
was left unchanged.
