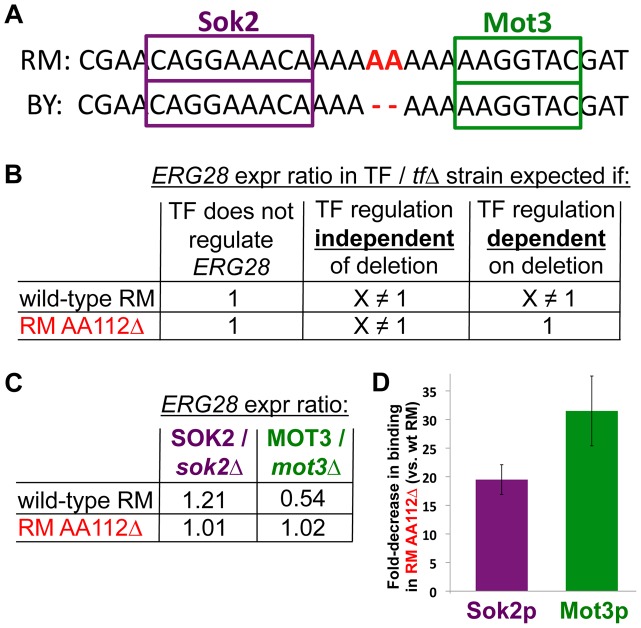
Figure 3. Determining the molecular mechanism of the causal mutation.
(A) Two predicted transcription factor (TF) binding sites flanking the deletion. (B) The expected fold-change in ERG28 expression level when deleting TFs under different scenarios. Left: if a TF does not regulate ERG28, its deletion should have no effect on ERG28 levels. Center: If a TF regulates ERG28 and acts independently of the two-base deletion, then deleting the TF should result in some fold-change X, which will be observed in both the wildtype RM and RM AA112Δ backgrounds. Right: If a TF regulates the wildtype ERG28 promoter, but the deletion abolishes this regulation, then the TF deletion may only affect ERG28 mRNA levels in the wildtype background (A fourth possible scenario, not shown, is where the TF only regulates ERG28 in RM AA112Δ). (C) qPCR data showing changes in ERG28 mRNA levels upon deleting either SOK2 or MOT3. In both cases, a difference is observed in the wildtype background (p = 7.5×10−5 for SOK2 and 5.0×10−3 for MOT3), but not the RM AA112Δ background (p = 0.28 for SOK2 and 0.67 for MOT3), consistent with the TF regulation being entirely abolished by the deletion. (D) Chromatin immunoprecipitation data showing the difference in binding for Sok2 and Mot3 to the ERG28 promoter in wildtype RM/RM AA112Δ. In both cases a significant decrease in binding is observed in RM AA112Δ.