Key Points
There is a role for the posttranslational modification, neddylation, in regulation of immune responses mediated by dendritic cells.
A role for neddylation in NF-κB signaling in dendritic cells was identified.
Abstract
Posttranslational protein modifications (PTMs) are necessary for cells to function properly. The role of PTMs in regulating immune responses, specifically those mediated by dendritic cells (DCs), which are critical for both innate and adaptive immunity, is not well understood. Utilizing multiple but complementary approaches, we determined the role of an important but less understood type of PTM, namely, neddylation, in regulating DC functions. Inhibition of neddylation suppressed the release of proinflammatory cytokines by DCs in response to Toll-like receptor, nucleotide oligomerization domain–like receptor, and noninfectious CD40L stimulation. These effects were more profound than those mediated by the proteasome inhibitor bortezomib or a commonly used antiinflammatory agent, dexamethasone. Targeting neddylation also suppressed the ability of DCs to stimulate murine allogeneic T cells in vitro and in vivo and human allogeneic T-cell responses in vitro. Mechanistic studies demonstrated that inhibition of neddylation reduced both canonical and noncanonical nuclear factor-κB (NF-κB) activity. Neddylation inhibition prevented the degradation of inhibitor-κB and thus reduced the translocation and activation of NF-κB, but without perturbation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway. Thus, blocking neddylation could be a novel strategy for mitigating immune-mediated disease processes.
Introduction
Posttranslational protein modifications (PTMs) are critical for immunity.1 PTMs include, but are not limited to, acetylation,2 phosphorylation, and ubiquitination.3 These PTMs are necessary for the proper function of all cells, including dendritic cells (DCs), which are critical for both innate and adaptive immunity.4-6 Following an encounter with inflammatory stimuli or components of microbial or viral origin, DCs upregulate costimulatory molecules7,8 and release proinflammatory cytokines.6 These proinflammatory cytokines play an important role in immunity in an autocrine and paracrine fashion on surrounding cells; however, the molecular processes that regulate cytokine release from DCs are not completely understood. Proinflammatory cytokines have profound impact on the causation or exacerbation of many disease states including chronic inflammatory diseases, shock, autoimmunity, and alloimmunity.9-12
Pattern recognition receptors (PRRs), such as Toll-like receptor (TLR) and nucleotide oligomerization domain (NOD)-like receptor (NLR), that are found on DCs interact with pathogen-associated molecular patterns (PAMPs), allowing selective recognition of microorganisms.13,14 A series of signaling events leads to the phosphorylation of inhibitor-κB (IκB) kinase β, which in turn phosphorylates IκB for degradation, resulting in the activation of the transcription factor nuclear factor-κB (NF-κB).15 At basal conditions, IκB sequesters canonical NF-κB in the cytosol. However, the critical molecular regulators of phosphorylated IκB degradation are not well understood. Activation and translocation of NF-κB result in the transcription of the proinflammatory cytokines, tumor necrosis factor-alpha (TNF-α) and interleukin-6 (IL-6).16 Activation of NF-κB occurs through 2 distinct canonical and noncanonical NF-κB pathways.16 NF-κB is comprised of heterodimers of the Rel protein family. The canonical NF-κB is a union of p65 (RelA) and p50 and responds to numerous PRR stimuli, resulting in diverse functions. In contrast, the noncanonical pathway consists of RelB and p52 constituting NF-κB and responds to a subset of TNFR signals such as CD40L.17
Upon PRR stimulation, IκB is phosphorylated and subsequently ubiquitinated, resulting in its degradation and allowing NF-κB to translocate to the nucleus.18 Phosphorylation of IκB is followed by its ubiquitination and degradation by the E3 ligase complex. This complex consists of the proteins Skp1 (S-phase kinase associated protein 1), an F-box protein, Cullin1 (Cul1), and a RING box protein (Rbx1 or Rbx2, also known as SAG), which together comprise the Cullin RING ligase-1 (CRL-1).19,20 The ability of CRL-1 to ubiquitinate target proteins, including IκB, is dependent upon the covalent PTM of the Cul1 subunit of the CRL by attachment of a ubiquitin homolog called NEDD8 (neural precursor cell expressed, developmentally downregulated 8).21 PTM by the attachment of NEDD8 is known as neddylation.22,23 Neddylation is an enzymatic process in which NEDD8 is activated in an ATP-dependent manner by an E1 enzyme known as NEDD8 activating enzyme (NAE) and is subsequently transferred to the E2 enzyme, Ubc12.24 NEDD8-Ubc12 transfers NEDD8 to Cullin, displacing CAND1, an inhibitor of Cullin activity.24,25 Once activated, CRL-1 catalyzes the transfer of ubiquitin from the E2 to the substrate.24 A small molecule called MLN4924, which has remarkable specificity and potency for inhibiting NAE, has been developed recently. MLN4924 forms a NEDD8-AMP mimetic and thereby prevents the initial activation of NEDD8.26 Currently, this small molecule is in human clinical trials as an anticancer agent; however, the role of neddylation in regulating immune cells, specifically DCs, is not known.
Utilizing 2 distinct but complementary approaches (chemical inhibition with the small molecule MLN4924 and molecular knockdown of critical CRL proteins), we examined the effect of inactivation of CRL-1 E3 ligase activity on murine and human DC function as determined by the release of proinflammatory cytokines and their ability to stimulate allogeneic T cells both in vitro and in vivo. We also compared these results with those for the known antiinflammatory drug dexamethasone and further determined the differential molecular mechanisms that underpin their antiinflammatory responses. We found that inactivation of the CRL-1 E3 ligase resulted in significantly greater suppression of proinflammatory cytokine production than dexamethasone or the proteasome inhibitor bortezomib. Also, mechanistic studies showed that in contrast to dexamethasone, inhibition of neddylation prevented the degradation of IκB as well as the translocation and activation of NF-κB without perturbation of the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) pathway.
Materials and methods
See supplemental Methods for details, available on the Blood website.
Reagents
RPMI1640, Dulbecco’s modified Eagle medium, alpha-minimum essential medium, penicillin, streptomycin, and sodium pyruvate were purchased from Gibco (Grand Island, NY); fetal calf serum from GemCell (Sacramento, CA); 2-mercaptoethanol from Sigma-Aldrich (St. Louis, MO); and murine granulocyte macrophage–colony-stimulating factor from Peprotech (Rocky Hill, NJ). All antibodies (Abs) used for the fluorescence-activated cell sorter were purchased from Biolegend (San Diego, CA). IL-6 enzyme-linked immunosorbent assay (ELISA) kits were purchased from BD Biosciences (San Diego, CA) and TNF-α, both mouse and human, from R&D Systems (Minneapolis, MN). Dimethylsulfoxide (DMSO) was obtained from Sigma-Aldrich, MLN4924 from Active BioChem (Maplewood, NJ), and dexamethasone from APP Pharmaceuticals (Schaumburg, IL); bortezomib was purchased from Fisher Scientific and lipopolysaccharide (LPS) from InvivoGen (San Diego, CA).
Mice
Female C57BL/6 (I-Ab; CD45.2+) and BALB/c (H2d) mice were purchased from the National Cancer Institute, Abb (B6.129-H2-Ab1tm1Gru N12) mice were purchased from Taconic, and bm12 (B6.C-H2-Ab1bm12/KhEg-Mc1re-J/J) mice were purchased from the The Jackson Laboratory. The age of mice used for experiments ranged from 7 to 12 weeks. All animals were cared for under regulations reviewed and approved by the University of Michigan’s Committee on Use and Care of Animals, based on University Laboratory Animal Medicine guidelines.
Isolation of bone marrow–derived DCs
DCs were obtained as described previously.27 See supplemental Methods for details. Studies with human cells were performed after obtaining informed consent from the participants; informed consent was obtained in accordance with the Declaration of Helsinki. The University of Michigan Institutional Review Board approved the studies (approval number: HUM00043287).
Cell culture
The DC cell line, JAWSII, was established from bone marrow (BM) cells of a p53-knockout C57BL/6 mouse and purchased from the American Type Culture Collection (CRL-11904; Manassas, VA). See supplemental Methods for details on culturing JAWSII cells.
Cytokine detection
Supernatants were collected and stored at −20°C until analysis. TNF-α and IL-6 ELISA kits (mouse and human; purchased from R&D Systems) were used per the manufacturer’s instructions and read at 450 nm using a SpectraMax microplate reader (Molecular Devices, Sunnyvale, CA).
Quantitative polymerase chain reaction
See supplemental Methods for details. Briefly, BM-derived DCs (BMDCs) were seeded on 60-mm plates with 3 × 106 cells per plate. Indicated cells were then pretreated for 2 hours with DMSO, MLN4924, or dexamethasone at indicated dosages. Cells were then stimulated with LPS (500 ng/mL) for 4 hours. Next, RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Following validation, quantitative polymerase chain reaction (qPCR) primers (see supplemental Methods) in conjunction with SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA) were used to quantify mRNA levels of cDNA (50 ng). qPCR was performed with a Mastercycler Realplex2 (Eppendorf, Hamburg, Germany) using the ΔΔCT method to calculate mRNA levels.
Flow cytometry
To analyze DC surface phenotype, DCs were incubated in the presence or absence of MLN4924 or vehicle (DMSO; Sigma-Aldrich). Cells were then harvested and stained with CD11c-conjugated allophycocyanin (APC) (clone: N418) and 1 of the following per triplicate group: Annexin V (BD Biosciences), CD80 (clone: 16-10A1), CD86 (clone: GL-1), major histocompatibility complex (MHC) II-I-Ab (clone: AF6-120.1), PD-L1 (clone: MIH5), PD-L2 (clone: TY25). All flow cytometry Abs were purchased from eBiosciences. Stained cells were then analyzed with an Accuri C6 flow cytometer (BD Biosciences).
Western blot and subcellular fractionation
See supplemental Methods for details. Briefly, equal amounts of protein from whole cell lysates were separated with sodium dodecyl sulfate–polyacrylamide gel electrophoresis and subsequently transferred to nitrocellulose membrane. See supplemental Methods for primary Abs used. Secondary Abs conjugated to horseradish peroxidase (Jackson ImmunoResearch) were used to detect primary Abs. Densitometric analysis was performed using ImageJ software. The cytoplasmic and nuclear fractions were isolated using the Nuclear Extract Kit (Active Motif) per the manufacturer’s instructions. The extracts where then analyzed via western blot.
Mixed lymphoctye reaction
Splenic T cells (2 × 105/well) were magnetically separated from WT-B6 or WT-BALB/c mice by autoMACS using CD90.2 microbeads and subsequently cultured with irradiated (30 Gy) WT-B6 DC at 40:1 (5 × 103/well) and at 100:1 (2 × 103/well), each for 72 and 96 hours. Human mixed lymphocyte reactions (MLRs) were performed by coculture of peripheral blood mononuclear cells (PBMCs; 1 × 105/well) and monocyte-derived dendritic cells at 1:1 (1 × 105) and at 10:1 (1 × 104) ratios, each for 96 and 120 hours. Incorporation of 3H-thymidine (1 µCi/well) by proliferating T cells or PBMCs during the final 6 hours of culture was measured by a TopCount (PerkinElmer).
Molecular knockdown
See supplemental Methods for details. Briefly, JAWSII cells were seeded and transfected with siRNA (3 µg) using oligofectamine reagent (Invitrogen) per the manufacturer’s instructions for 24 hours before stimulation with LPS. RNA was extracted from cell pellets using an RNeasy Mini Kit (Qiagen) and analyzed via qPCR.
Confocal microscopy
See supplemental Methods for details. BMDCs were seeded onto Corning glass coverslips (1 × 105 cells/slip; Fisher Scientific) overnight at 37°C. Cover slips were then washed, fixed with 4% paraformaldehyde for 20 minutes, and subsequently permeabilized with 0.3% Triton X. Cells were stained with NF-κB p65 primary and Alexa Fluor 488 secondary, 4,6 diamidino-2-phenylindole (DAPI; Invitrogen), and Alexa Fluor 555 phalloidin. Coverslips were then mounted using ProLong Gold antifade reagent (Molecular Probes), and Z-stack images were acquired at room temperature using a Nikon A-1 confocal microscope (Mellville, NY) equipped with an oil immersion 60× objective with a numerical aperture equal to 1.4 and imported into NIS-Elements Software (Nikon). Excitation lasers of 405 nm, 488 nm, and 561 nm were used.
Statistical analyses
All statistical analyses were performed using GraphPad Prism (GraphPad Software). P values were calculated using the Student's t test.
Results
Inhibition of neddylation attenuates LPS-induced proinflammatory cytokine production by DCs
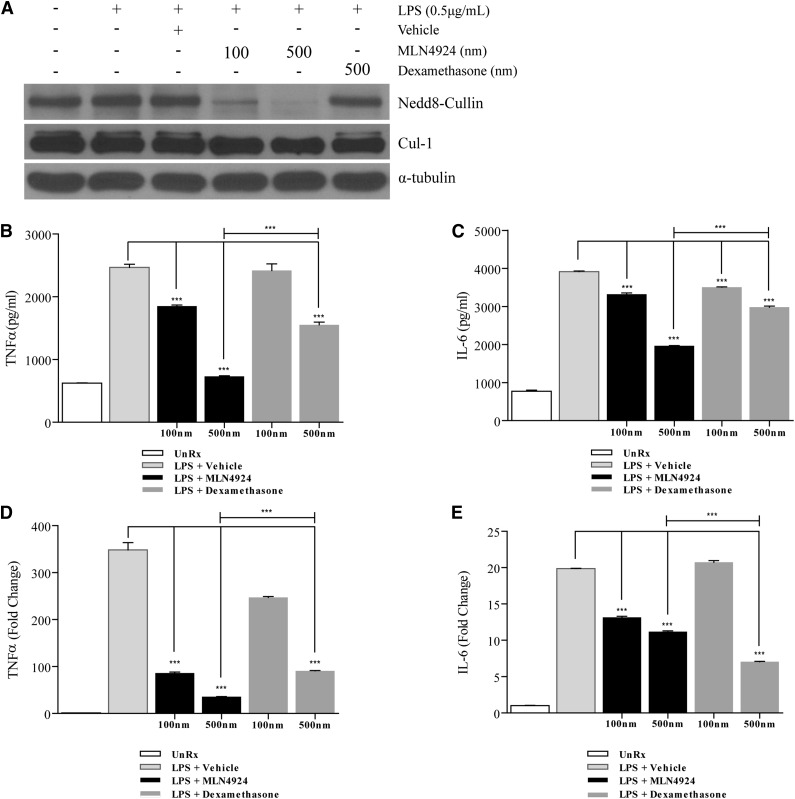
Previous studies have shown that MLN4924 prevents the neddylation of CRLs in several cell types.28,29 Therefore, we determined whether MLN4924 or dexamethasone could inhibit neddylation in BMDCs. BMDCs that were pretreated with MLN4924 followed by TLR4 stimulation (LPS) exhibited reduced neddylation (Figure 1A). This inhibition of neddylation was detected using a NEDD8 Ab capable of binding NEDD8-conjugated Cullin protein at approximately 90 kDa (Figure 1A, top panel).28,30 We further confirmed this with an Ab specific for total Cul119 (Figure 1A, middle panel). By contrast, treatment with dexamethasone did not inhibit neddylation in BMDCs (Figure 1A).
Figure 1.
Neddylation inhibition attenuates LPS-induced TNF-α and IL-6 release and gene expression in BMDC. Protein analysis by western blot of (A, top panel) neddylated Cullin proteins and (A, middle panel) Cul1 protein using whole cell lysate of BMDC stimulated with LPS in the presence or absence of MLN4924 or dexamethasone at the indicated doses. Cells receiving treatment were preincubated with MLN4924 or dexamethasone for 2 hours followed by concurrent LPS stimulation (0.5 µg/mL) for 4 hours. α-tubulin protein levels were analyzed as an equal loading control. Quantification via ELISA of cytokines TNF-α (B) and IL-6 (C) and qPCR analysis of TNF-α (D) and IL-6 (E) transcripts in BMDC stimulated with LPS in the presence or absence of MLN4924 or dexamethasone at the indicated doses. Cells receiving treatment were preincubated with MLN4924 or dexamethasone for 2 hours followed by concurrent LPS stimulation (0.5 µg/mL) for 4 hours. One representative experiment of 3 is shown. ***P < .0001.
Having established that MLN4924 inhibited neddylation, next we determined whether it modulated the production of proinflammatory cytokines such as TNF-α and IL-6 following stimulation of BMDCs with LPS.31 TNF-α (Figure 1B) and IL-6 (Figure 1C) cytokine levels in the supernatants were significantly inhibited by MLN4924 in a dose-dependent manner. Furthermore, MLN4924 suppressed LPS-induced inflammatory cytokines significantly more than dexamethasone treatment (Figure 1B-C). Because LPS increases the expression of TNF-α and IL-6 at the transcriptional level,32,33 we assessed the impact of MLN4924 and dexamethasone on LPS-induced transcriptional expression of TNF-α and IL-6 mRNA. As shown in Figure 1D-E, both MLN4924 and dexamethasone inhibited LPS-induced TNF-α and IL-6 mRNA levels. The impact of MLN4924 on mitigating LPS-induced increases of TNF-α and IL-6 mRNA was dose dependent (Figure 1D-E).
Collectively, these data suggest that MLN4924 and dexamethasone differentially affect Cul1 neddylation in DCs and that they may not have interrelated molecular targets. More importantly, the data show that inhibition of neddylation results in a more significant attenuation of LPS-induced TNF-α and IL-6 production than dexamethasone in BMDC.
Neddylation modulates DC responses to both infectious and noninfectious stimuli
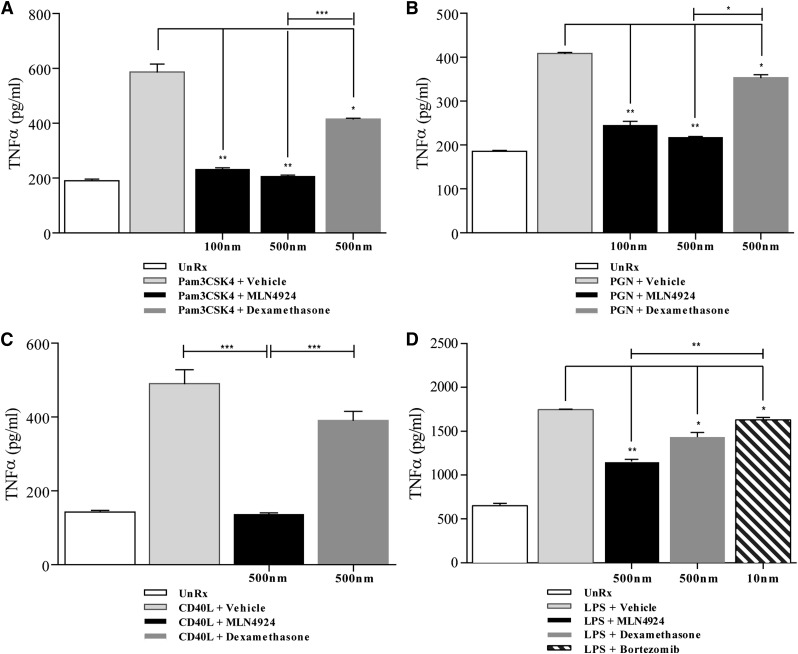
Next, in order to determine whether the effects of MLN4924 observed thus far were specific only to TLR4 ligation (LPS), we stimulated BMDC with Pam3CSK4 (a TLR1/2 ligand), peptidoglycan (PGN; a NOD2 stimulator), and noninfectious CD40L agonist. Treatment of DCs in the presence or absence of MLN4924 or dexamethasone caused a significant reduction in the secretion of TNF-α by both MLN4924 and dexamethasone, regardless of whether they were stimulated with Pam3CSK4 (Figure 2A), PGN (Figure 2B), or CD40L (Figure 2C). These data suggest that the effects on suppression of proinflammatory cytokines through inactivation of the CRL-1 E3 ligase are not specific to TLR4 but extend to an array of PRR and cellular stimuli such as CD40L. Furthermore, these data also show that inhibition of neddylation regulated both canonical (TLR/NLR) and noncanonical (CD40L) activated NF-κB pathways. To further compare the effects of neddylation inhibition with another clinically used inhibitor of the proteasome and NF-κB pathway, we stimulated BMDC with LPS and concurrently treated with the diluent vehicle, MLN4924, dexamethasone, or bortezomib. As shown in Figure 2D, blocking neddylation inhibited TNF-α secretion from the DCs far more potently than the proteasome inhibitor bortezomib.
Figure 2.
Neddylation inhibition in BMDC mitigates release of non-TLR4–stimulated cytokines in vitro. ELISA quantification of TNF-α release in Pam3CSK4 (300 ng/mL) stimulated (A), PGN (5 μg/mL) stimulated (B), and CD40L (1 μg/mL) stimulated (C) BMDC in the presence or absence of vehicle, MLN4924, or dexamethasone at the indicated doses. (D) Comparison of TNF-α release via ELISA of cells treated with vehicle, MLN4924, dexamethasone, or bortezomib followed by concurrent stimulation with LPS (0.5 µg/mL). Cells receiving treatment were preincubated with vehicle, MLN4924, dexamethasone, or bortezomib for 2 hours followed by concurrent stimulation for 4 hours. One representative experiment of 3 is shown in cells treated in triplicate. *P < .05; **P < .01; ***P < .0001.
Inhibition of neddylation mitigates the ability of DCs to induce proliferation of allogeneic T cells
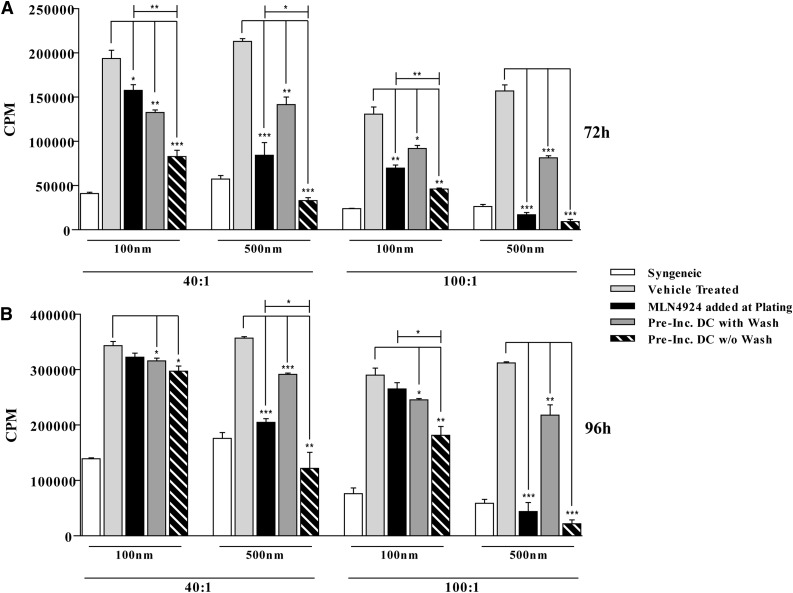
To further characterize the effect of neddylation inhibition on the function of DCs, we determined the impact of treating DCs with MLN4924 on their ability to stimulate allogeneic T cells4,5 in a MLR. BALB/C T cells were cultured with C57BL/6 irradiated (30 Gy) BMDC at 40:1 and 100:1 ratios for 72 hours (Figure 3A) and 96 hours (Figure 3B). Addition of MLN4924 to the cultures upon plating cells significantly reduced T-cell proliferation (Figure 3A-B). To determine whether this reduction was due to a direct impact of MLN4924 on DCs, BMDCs were preincubated with MLN4924 for 6 hours, washed, and then used as stimulators in the MLR. Pretreatment of DCs with MLN4924 also reduced proliferation of allo-T cells at both 72 hours (Figure 3A) and 96 hours (Figure 3B), suggesting that MLN4924 treatment of DCs reduced their ability to induce proliferation of allo-T cells. To specifically determine whether inhibition of neddylation can also suppress T cells directly, we stimulated BALB/C CD90.2+ T cells with α-CD3 and α-CD28 functional Abs for 48 hours following their pretreatment in the presence or absence of MLN4924. MLN4924 significantly reduced T-cell proliferation (supplemental Figure 3A) and caused a significant decrease in the secretion of interferon-γ (supplemental Figure 3B).
Figure 3.
Neddylation inhibition mitigates allogeneic T-cell proliferation. BALB/C T cells were cultured with C57BL/6 irradiated (30 Gy) BMDC at 40:1 and 100:1 ratios in the presence or absence of MLN4924 for 72 hours (A) and 96 hours (B). Where indicated, BMDC were preincubated in the presence or absence of vehicle or MLN4924 at specified dosage for 6 hours. One representative experiment of 3 is shown of groups treated in triplicate. *P < .05; **P < .01; ***P < .0001.
Inhibition of neddylation regulates DC functions in vivo
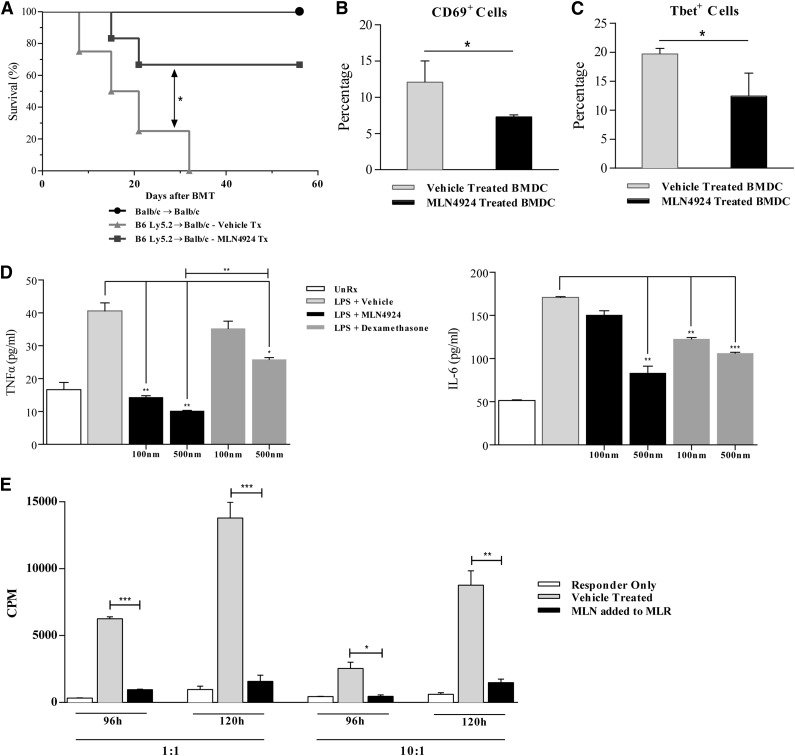
Next we investigated the effects of neddylation inhibition on in vivo DC functions. Because host antigen–presenting cells modulate graft-vs-host disease (GVHD) and DCs are the most potent APCs, we used a clinically relevant, MHC-mismatched B6 (H-2b) → BALB/C (H-2d) model of allogeneic bone marrow transplant (BMT) to test the effect of neddylation inhibition on induction and severity of GVHD. Recipient BALB/C animals were lethally irradiated with 8 Gy on day −1 and transplanted with 0.5 × 106 CD90.2+ T cells and 5 × 106 BM cells from either syngeneic BALB/C or allogeneic MHC-mismatched C57BL/6 donors on day 0. The recipients also received either MLN4924 (20 mg/kg) or vehicle from day −1 to day 3 because host DCs typically cannot be recovered beyond this time following allo-BMT. All syngeneic recipients survived with no signs of GVHD, demonstrating absence of nonspecific toxicity by MLN4924 (Figure 4A). The allogeneic recipients that received only vehicle showed signs of severe GVHD and greater mortality than syngeneic animals (Figure 4A). By contrast, allogeneic recipients receiving MLN4924 exhibited significantly improved survival compared with vehicle control (Figure 4A), suggesting that in vivo blockade of neddylation mitigated GVHD.
Figure 4.
Neddylation blockade regulates DC-mediated T-cell activation. (A) Survival of BALB/C animals lethally irradiated and transplanted with BM and CD90.2+ T cells from either syngeneic BALB/C or allogeneic C57BL/6 donors. Following lethal irradiation (8 Gy) on day 1, MLN4924 (20 mg/kg) was administered in 5 daily doses, day 1 to day 3 relative to BMT. Data are combined from 2 independent experiments (n = 10 to 12 animals in the allogeneic groups). (B-C) Abb animals were lethally irradiated (10 Gy) on day 1 and WT B6 BMDC (10 × 106) pretreated with vehicle or MLN4924 overnight and subsequently transferred in 2 doses separated by 24 hours (day 1 and day 0). CD90.2+ T cells (2 × 106) from syngeneic WT-C57BL/6 or allogeneic bm12 animals were transferred on day 0. Following sacrifice on day 6, spleens were analyzed for CD69 (B) and Tbet (C). (D) Quantification via ELISA of TNF-α (left) and IL-6 (right) released from human moDCs stimulated with LPS in the presence or absence of MLN4924 or dexamethasone at the indicated doses. Cells receiving treatment were preincubated with vehicle, MLN4924, or dexamethasone for 2 hours followed by concurrent LPS stimulation (0.5 µg/mL) for 4 hours. One representative experiment of 3 is shown of groups treated in triplicate. (E) Human PBMCs (1 × 105/well) and moDCs were obtained from 2 healthy donors and cocultured at 1:1 and 10:1 ratios in an MLR in the presence or absence of MLN4924 for 96 and 120 hours.
Because systemic administration of MLN4924 is likely to impact not just DCs but also several other cells, particularly donor T cells that are critical for GVHD,34 the reduction in GVHD mortality might be a reflection of its effects on these other cells in vivo. Therefore, in order to evaluate the effects of neddylation inhibition only on the in vivo function of BMDCs without the confounding effects on other tissues or expression of target antigens, we devised a model in which allogeneic CD4+ T cells would respond only to MHC class II alloantigens on exogenously administered DCs in an acute GVHD model. MHC class II–deficient (Abb [H2-Ab1]) C57BL/6 mice (H2b)8 received 10 Gy total body irradiation and were injected with 1 × 107 BMDCs from syngeneic WT C57BL/6 (H2b) animals that were incubated overnight with either vehicle or MLN4924 in 2 doses separated by 24 hours. We then injected 2 × 106 CD90.2+ T cells from either syngeneic C57BL/6 or allogeneic bm12 donors (see Materials and methods and supplemental Figure 2), which differ from the recipient animals by a single MHC class II antigen. Analysis of donor T cells in the spleen at day 6 revealed fewer activated CD69+ (Figure 4B) and differentiated Tbet+ (Figure 4C) CD4+ T cells in animals that received DCs pretreated with MLN4924.
Blockade of neddylation attenuates the functions of human DCs
To determine the clinical relevance of our observations, we determined whether the effects of neddylation inhibition on DCs was also germane to DCs derived from healthy human PBMCs. Human peripheral blood moDCs were harvested and cultured with and without MLN4924 and stimulated with LPS. The secretion of TNF-α and IL-6 by LPS from moDCs,35 were significantly reduced by MLN4924 compared with vehicle control (Figure 4D). Similar to the effect on murine DCs, the reduction of proinflammatory cytokines was also significantly greater than dexamethasone (Figure 4D).
Next, we determined whether MLN4924 attenuated the ability of human monocyte-derived DCs to stimulate allogeneic PBMCs. PBMCs and moDCs were obtained from 2 healthy donors and cultured in an MLR at 1:1 and 10:1 ratios in the presence or absence of MLN4924. Following culture for 96 hours and 120 hours, a significant decrease in proliferation of PBMCs was observed in cultures incubated with MLN4924 (Figure 4E), similar to the results observed in murine cells.
Inhibition of neddylation does not increase apoptosis or alter the phenotype of DCs
Studies have shown that MLN4924 reduces the viability of cancer cell lines.29 Therefore, we determined whether the observed effects of neddylation inhibition through inactivation of Cul1 E3 ligase by MLN4924 was due to reduced BMDC viability. Treatment of BMDCs with MLN4924 for 24 hours at the doses that reduced their function did not cause greater apoptosis and thus did not affect cell viability (supplemental Figure 1B). Next, we assessed whether MLN4924 arrested the maturation of DCs as determined by the expression of surface costimulatory molecules. To determine this, we supplemented the culture media during BMDC generation with vehicle or MLN4924 for the last 24 hours and then examined cell surface expression of CD80, CD86, MHC II, PD-L1, and PD-L2 with flow cytometry. Addition of MLN4924 did not cause phenotypic changes (supplemental Figure 1A). Taken together, these data suggest that the reduction of DC function following treatment with MLN4924 is not due to the result of decreased cell viability or phenotypic change, but that these are intrinsic to inhibition of neddylation.
Inhibition of SAG reduces LPS-induced cytokine production in DCs
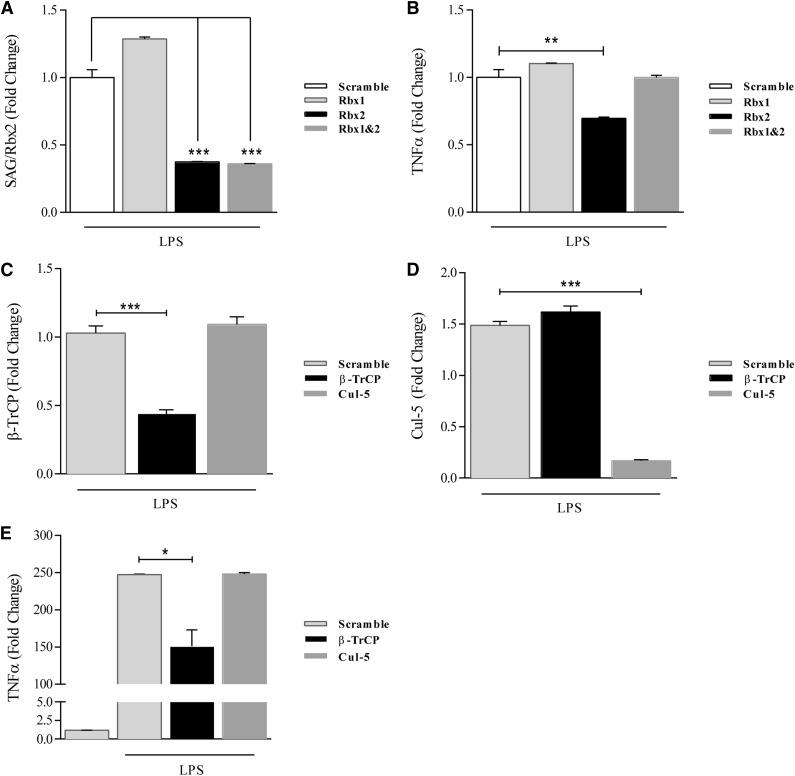
The inhibition of LPS-induced cytokine production by MLN4924 suggests that deneddylation is critical to this process by inactivation of CRL-1 E3 ligase activity. To determine the specificity of the approach and to rule out any potential off-target effects of MLN4924, we used siRNA-mediated knockdown of RBX2/SAG, a critical and specific component of the CRL-1–induced neddylation pathway. Next we performed siRNA-mediated knockdown of RBX1 and RBX2/SAG alone and in combination in JAWSII cells. First, we confirmed the efficiency and specificity of knockdown of targeting SAG transcripts with siRNA, as indicated by reduced levels of SAG (Figure 5A). Then we compared TNF-α cytokine release following LPS stimulation in JAWSII cells transfected with the control-scrambled siRNA or SAG-specific siRNA. Upon LPS stimulation, JAWSII cells transfected with SAG siRNA exhibited a significant reduction in expression of TNF-α compared with the scrambled siRNA (Figure 5B). Interestingly, double knockdown of Rbx1 and SAG/Rbx2 had little or no effect on cytokine production, which could be because Rbx1 might antagonize or cause feedback inhibition of SAG (Rbx2). Future studies will address this issue. Nonetheless, collectively, these data suggest that inhibition of neddylation through inactivation of the CRL-1 mediated either by siRNA knockdown of SAG or by the small molecule MLN4924 reduced proinflammatory cytokine release by DCs.
Figure 5.
siRNA-mediated neddylation inhibition inhibits LPS-induced TNF-α production in JAWSII cells. (A) qPCR analysis of SAG expression in JAWSII cells transfected with indicated siRNA (3 μg). Scramble transfection performed as control. (B) qPCR analysis of TNF-α expression in JAWSII cells transfected with indicated siRNA and stimulated with LPS (0.5 µg/mL). Scramble transfection performed as control. One representative experiment of 3 is shown. (C-E) JAWSII cells transfected with siRNA for βTrCP, Cul5, or scramble as described in Materials and methods. Expression of βTrCP (C) and Cul5 (D) mRNA transcripts in JAWSII cells receiving indicated siRNA-mediated knockdown. (E) Expression of TNF-α mRNA transcripts in cells transfected with indicated siRNA and cultured in the presence or absence of LPS (0.5 µg/mL) for 4 hours. *P < .05; **P < .01; ***P < .0001.
Inhibition of βTrCP but not Cul5 reduces LPS-induced cytokine expression in DCs
Inhibition of neddylation via MLN4924 and molecular knockdown of Rbx2 results in global CRL inactivation. Therefore, to determine if the observed results are specific to CRL-1, we silenced beta-transducin repeat-containing protein (β-TrCP) as well as Cul5 using siRNA-mediated knockdown in JAWSII cells. First, we ensured efficient knockdown of both βTrCP (Figure 5C) and Cul5 (Figure 5D). Next, we examined the expression of TNF-α as a functional readout from the cells transfected with scramble, βTrCP, or Cul5 and subsequently stimulated with LPS. A significant decrease in TNF-α expression was observed in cells receiving knockdown of βTrCP, but not in cells with Cul5 knockdown (Figure 5E). These data suggest that the function of CRL-1 is critical for the underlying mechanism of immune suppression following inhibition of neddylation.
MLN4924 inhibits translocation and activation of NF-κB
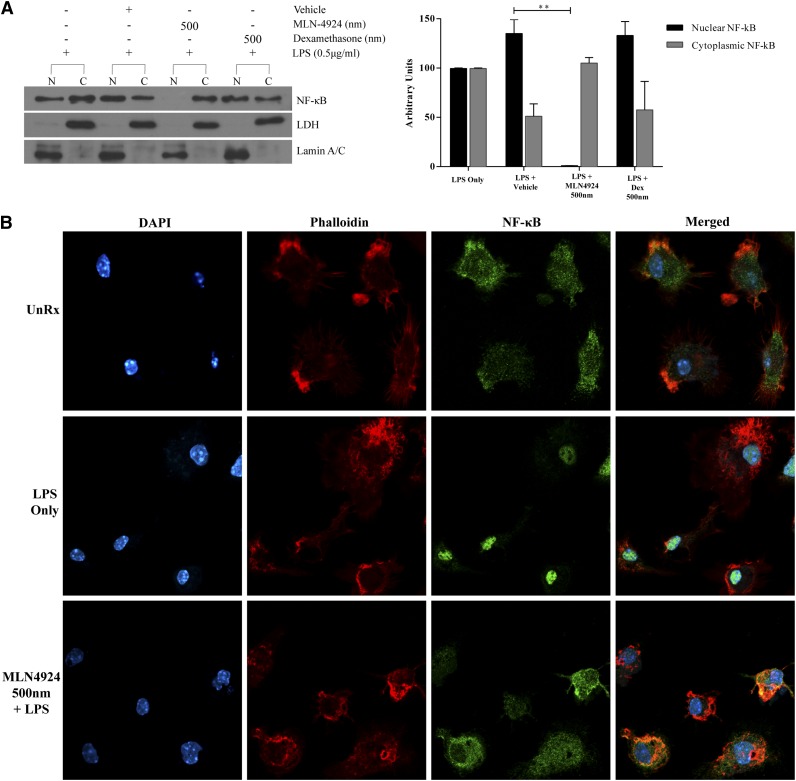
To determine if neddylation inhibition prevents transcriptional activity of LPS-induced NF-κB, we assessed p65 NF-κB translocation to the nucleus in BMDCs treated with MLN4924 or dexamethasone following stimulation with LPS. As anticipated, cells cultured in the presence of vehicle and stimulated with LPS for 30 minutes, p65 NF-κB was present in the nuclear fraction (Figure 6A, left panel). Cells pretreated with 500 nm dexamethasone and stimulated with LPS exhibited a similar level of p65 NF-κB in the nuclear fraction (Figure 6A, left, top panel). However, pretreating cells with MLN4924 prior to LPS stimulation inhibited protein levels of p65 NF-κB in the nuclear fraction of BMDCs (Figure 6A). Interestingly, these doses of MLN4924 and dexamethasone inhibited both LPS-induced release and gene expression of TNF-α and IL-6 (Figure 1B-E). These data indicate that LPS-induced nuclear accumulation of p65 NF-κB in BMDCs is blocked by MLN4924 but not dexamethasone, suggesting that MLN4924 and dexamethasone have divergent molecular effects.
Figure 6.
Neddylation inhibition and dexamethasone differentially affect NF-κB translocation in BMDC. (A) Protein analysis by western blot (left) of p65 isoform of NF-κB protein using nuclear (N) and cytosolic (C) fractions of BMDC in the presence or absence of 500 nm MLN4924 or 500 nm dexamethasone and stimulated concurrently with LPS (0.5 µg/mL) for 30 minutes. Plot (right) shows densitometric and statistical analysis of p65 NF-κB presence in the nuclear fraction and cytosolic fraction. Lamin A/C and LDH proteins were analyzed to demonstrate the presence of nuclear and cytosolic fractions, respectively. One representative experiment of 3 is shown. (B) Immunocytochemistry analysis of p65 NF-κB (column 3) localization in BMDC cultured in the presence or absence of MLN4924 and stimulated with LPS (0.5 µg/mL) for 1 hour. Cell nuclei were stained using DAPI (column 1). Cytoplasmic actin was stained with phalloidin (column 2). Merged images of staining (column 4). One representative experiment of 3 is shown. **P < .01.
Next, in order to visualize and demonstrate the translocation of p65, we performed immunocytochemistry. BMDC cultured only with vehicle exhibited a diffuse cytoplasmic localization of NF-κB (Figure 6B, top row). By contrast, BMDCs stimulated with LPS for 1 hour and treated with vehicle showed prominent NF-κB accumulation in the nucleus (Figure 6B, middle row). However, LPS-stimulated cells treated with MLN4924 showed a dispersed cytoplasmic localization of NF-κB similar to unstimulated controls (Figure 6B, bottom row). These complementary methods of examining NF-κB translocation showed prevention of translocation of NF-κB to the nuclei in BMDCs treated with MLN4924.
Inhibition of neddylation prevents IκB degradation in BMDC
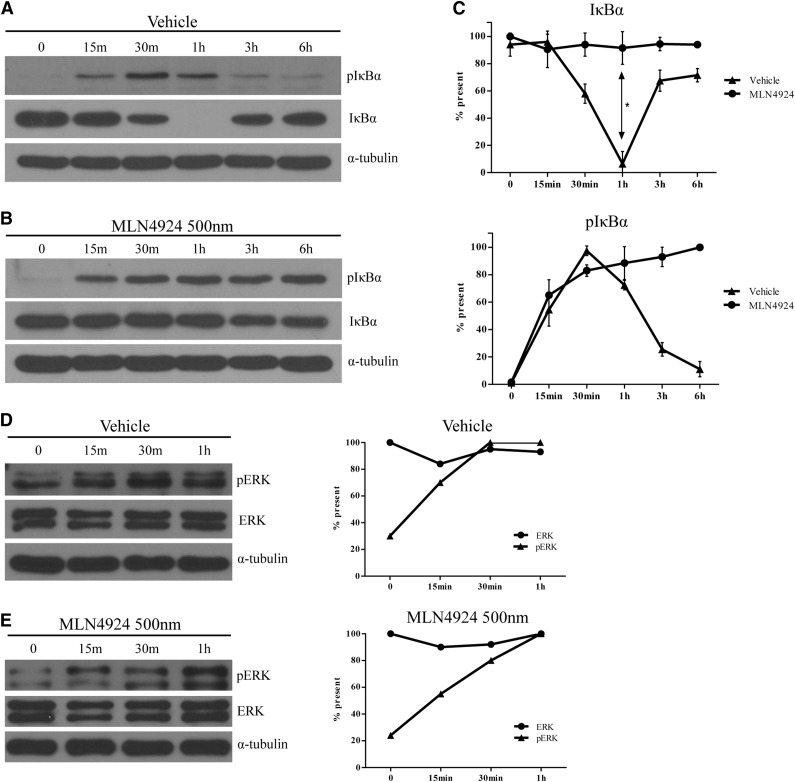
Previous studies have shown that IκB is a target of CRLs.36 Therefore, we examined the effect of MLN4924 on the degradation of IκB in BMDCs when stimulated with LPS. Similar to previous studies of BMDCs37 and other APCs,38 stimulation with LPS resulted in the reduction of IκB protein levels within 30 minutes in vehicle-treated BMDCs. Levels of IκB were slowly restored over a 6-hour period (Figure 7A, middle panel). The observed decrease in IκB following LPS stimulation coincided with increased phosphorylation of IκB at serine-32 and serine-36 during the first 30 minutes of treatment. The increased p-IκB slowly returned to lower levels during the remaining 5 hours of treatment (Figure 7A, top panel). Treatment of BMDCs with 500 nM MLN4924 inhibited any LPS-mediated decrease in IκB (Figure 7B, middle panel), while levels of phosphorylated IκB in the same cells were elevated within 15 minutes following LPS treatment (Figure 7B, top panel). Further, the levels of phosphorylated IκB remained elevated for the duration of the 6-hour treatment (Figure 7B). Following densitometric and statistical analyses, the decrease in IκB in vehicle-treated cells was significantly greater than in MLN4924-treated cells (Figure 7C, top panel). These data indicate that inhibition of neddylation blocks the degradation of phosphorylated IκBSer32/Ser36 protein in BMDCs.
Figure 7.
Neddylation inhibition prevents IκB degradation in BMDC without perturbation of the MAPK/ERK pathway. Protein analysis via western blot of phosphorylated IκBα protein (A-B, top panels) and total IκBα (A-B, middle panels) using whole cell lysate of BMDC stimulated with LPS (0.5 µg/mL) for 6 hours in the presence of vehicle (A) or 500 nm MLN4924 (B). Densitometric and statistical analysis of total IκB (C, top) or pIκB (C, bottom) of vehicle or MLN4924-treated cells. α-tubulin protein levels were analyzed as an equal loading control. One representative experiment of 3 is shown. Protein analysis via western blot of phosphorylated ERK protein (D-E, top panels) and total ERK (D-E, middle panels) using whole cell lysate of BMDC stimulated with LPS for 6 hours in the presence of vehicle (D) or 500 nm MLN4924 (E). Densitometric analysis of vehicle-treated cells (D, right) or 500 nm MLN4924-treated cells (E, right) of phosphorylated ERK and total ERK protein. α-tubulin protein levels were analyzed as an equal loading control. One representative experiment of 3 is shown. *P < .05.
Neddylation blockade does not perturb LPS-induced activation of MAPK/ERK pathway
Previous studies have shown that LPS stimulation of cells results in activation of the MAPK/ERK pathway.37 Therefore, we examined whole cell lysates of BMDC stimulated with LPS cultured in the presence of vehicle or MLN4924. Western blot analysis showed that ERK protein levels were similar in vehicle-treated cells (Figure 7D) and MLN4924-treated cells (Figure 7E). In addition, levels of pERK increased in a similar manner in both vehicle- and MLN4924-treated cells (Figure 7D-E). These data suggest that MLN4924 treatment specifically reduced proinflammatory cytokines through regulation of the NF-κB pathway without affecting the MAPK/ERK pathway.
Discussion
PTMs modulate immunity.1 Proteins critical for an effective immune response, such as NF-κB, are known to be phosphorylated and acetylated.39,40 However, the role that neddylation plays in regulating immune cells, such as DCs, through the translocation of NF-κB is not clear. Our results demonstrate a novel role for PTM neddylation in the regulation of DC-mediated immunity. The mechanism of neddylation characterizes a novel molecular target for the inhibition of secretion and transcription of certain LPS-induced proinflammatory cytokines in DCs. We show that following inhibition of neddylation and stimulation via TLR and NLR, release and gene expression of the proinflammatory cytokines TNF-α and IL-6 are greatly mitigated (Figures 1B-E and 2).
Studies have shown the ability of MLN4924 to suppress the growth of tumor cell lines as well as primary human acute myeloid leukemia cells.19,22,30 Inhibition of the NEDD8 pathway by MLN4924 has been reported to result in apoptosis.26,41 However, our data suggest that MLN4924 did not affect BMDC viability (supplemental Figure 1B), demonstrating that the reduction in the production of proinflammatory cytokines was not due to loss of cells but was a functional consequence of neddylation inhibition. Our data also suggest that treatment with MLN4924 had a significantly greater impact on suppression of the release of proinflammatory cytokines than dexamethasone or bortezomib (Figures 1 and 2) in the nanomolar dose range. Moreover, inhibition of neddylation with MLN4924 reduced DC functions in vivo and attenuated GVHD (Figure 4A-C), suggesting that this pathway could be targeted to mitigate in vivo inflammatory diseases.
Mechanistic studies have uncovered a distinct molecular pathway for MLN4924-mediated suppression of DC functions when compared with dexamethasone. The primary molecular target of dexamethasone is the glucocorticoid receptor.42 Binding of steroid results in activation of the receptor and subsequent translocation to the nucleus43 and is known to interact with transcription factors such as NF-κB and AP-1.44 These interactions repress the expression of genes that code a number of cytokines that play key roles in the immune and inflammatory systems. By contrast, MLN4924 specifically targets NAE, a heterodimer of NAE1 and UBA3 subunits.26 By doing so, MLN4924 inactivates CRLs and impedes the ubiquitination rate of the subset of proteins whose degradation is dependent on CRLs.26
Although dexamethasone significantly inhibited secretion of TNF-α and IL-6, which are known to be regulated by NF-κB,16 it did not directly prevent the translocation of NF-κB to the nucleus after LPS stimulation (Figure 6A), in contrast to MLN4924. The ability of MLN4924 to directly prevent the degradation of IκB in LPS-stimulated cells (Figure 7B) suggests a specific and distinct mechanism from that of dexamethasone. The role of inhibiting neddylation, by MLN4924, in suppressing the release of TNF-α and IL-6 through the inactivation of CRL-1 causing IκBα accumulation and subsequent prevention of NF-κB activation, is further supported by molecular inactivation of CRL-1 by the siRNA knockdown of SAG and βTrCP (Figure 5). Together, these data suggest that MLN4924 and dexamethasone block LPS-induced cytokine release through distinct mechanisms.
The results of our study are in agreement with previous reports in cell lines demonstrating an effect on NF-κB activation.45 For the first time, we extend those observations in primary cells, DCs, and their functional immune responses. More importantly, we show a similar effect on primary human DCs. Furthermore, we uncover the molecular mechanism that is dependent on IκB degradation but independent of the MAPK/ERK pathway (Figure 7) and show that it is germane to both canonical and noncanonical NF-κB signaling-mediated release of proinflammatory cytokines. Other studies have shown that degradation of IκB and release of proinflammatory cytokines can be altered by proteasome inhibitors.46 However, these proteasome inhibitors also activate the transcription factor AP-1, which is known to contribute to LPS-induced cytokines.37 Our results show that upon treatment of LPS-stimulated BMDC with MLN4924, the MAPK/ERK pathway is unperturbed (Figure 7D-E), suggesting that AP-1 activity is unaltered by neddylation inhibition. This is likely due to the degradation of IL-1 receptor-associated kinase 1 through its association with the E3 ligases TNF receptor–associated factor 6 and Pellino.47 Previous studies have shown that Pellino, not CRL, is a likely candidate to serve as the degradation scaffold for IL-1 receptor-associated kinase 1.48 Thus, inhibition of the neddylation pathway may be a more specific approach of inhibiting the degradation of IκB and thereby blocking the increase in proinflammatory cytokines as compared with the effects seen by proteasome inhibitors.
Promising preclinical results led to the advancement of MLN4924 to phase 1 clinical trials for both nonhematological and hematological cancers.49,50 However, MLN4924’s immune-modulatory effects have heretofore been largely unrecognized. Our results suggest that at noncytotoxic doses, drugs such as MLN4924 that specifically target neddylation might provide novel therapeutic strategies for diseases associated with deregulated immune responses such as GVHD, chronic inflammatory disease, shock, autoimmunity, and allotransplantation.9-12,29
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants (National Heart, Lung and Blood Institute, HL-090775; National Cancer Institute, CA143379; and National Institute of Allergy and Infectious Diseases, AI-075284).
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: N.M. designed and performed experiments, analyzed data, and wrote the paper; T.T. designed and performed experiments and analyzed data; S.K., Y.S., K.O.-W., H.T., Y.W. and G.H. performed experiments; Y.S. analyzed data, contributed reagents, and assisted with writing the paper; and P.R. designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pavan Reddy, Department of Internal Medicine, Division of Hematology and Oncology, Blood and Marrow Transplantation Program, University of Michigan Comprehensive Cancer Center, 3312 CCGC, 1500 E Medical Center Dr, Ann Arbor, MI, 48105-1942; e-mail: reddypr@med.umich.edu.
References
- 1.Cloos PAC, Christgau S. Post-translational modifications of proteins: implications for aging, antigen recognition, and autoimmunity. Biogerontology. 2004;5(3):139–158. doi: 10.1023/B:BGEN.0000031152.31352.8b. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Chin YE, Weisiger E, et al. Cutting edge: Negative regulation of dendritic cells through acetylation of the nonhistone protein STAT-3. J Immunol. 2009;182(10):5899–5903. doi: 10.4049/jimmunol.0804388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magnani M, Crinelli R, Bianchi M, Antonelli A. The ubiquitin-dependent proteolytic system and other potential targets for the modulation of nuclear factor-kB (NF-kB). Curr Drug Targets. 2000;1(4):387–399. doi: 10.2174/1389450003349056. [DOI] [PubMed] [Google Scholar]
- 4.Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol. 2012;13(12):1145–1154. doi: 10.1038/ni.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hespel C, Moser M. Role of inflammatory dendritic cells in innate and adaptive immunity. Eur J Immunol. 2012;42(10):2535–2543. doi: 10.1002/eji.201242480. [DOI] [PubMed] [Google Scholar]
- 6.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106(3):255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 7.Turley SJ, Inaba K, Garrett WS, Ebersold M, Unternaehrer J, Steinman RM, Mellman I. Transport of peptide-MHC class II complexes in developing dendritic cells. Science. 2000;288(5465):522–527. doi: 10.1126/science.288.5465.522. [DOI] [PubMed] [Google Scholar]
- 8.Grusby MJ, Johnson RS, Papaioannou VE, Glimcher LH. Depletion of CD4+ T cells in major histocompatibility complex class II-deficient mice. Science. 1991;253(5026):1417–1420. doi: 10.1126/science.1910207. [DOI] [PubMed] [Google Scholar]
- 9.Tabas I, Glass CK. Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science. 2013;339(6116):166–172. doi: 10.1126/science.1230720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin GS. Sepsis, severe sepsis and septic shock: changes in incidence, pathogens and outcomes. Expert Rev Anti Infect Ther. 2012;10(6):701–706. doi: 10.1586/eri.12.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J. Adipose tissue macrophages in the development of obesity-induced inflammation, insulin resistance and type 2 Diabetes. Arch Pharm Res. 2013;36(2):208–222. doi: 10.1007/s12272-013-0023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tawara I, Koyama M, Liu C, et al. Interleukin-6 modulates graft-versus-host responses after experimental allogeneic bone marrow transplantation. Clin Cancer Res. 2011;17(1):77–88. doi: 10.1158/1078-0432.CCR-10-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6(9):644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 14.Shaw MH, Reimer T, Kim Y-G, Nuñez G. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr Opin Immunol. 2008;20(4):377–382. doi: 10.1016/j.coi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moynagh PN. The Pellino family: IRAK E3 ligases with emerging roles in innate immune signalling. Trends Immunol. 2009;30(1):33–42. doi: 10.1016/j.it.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Razani B, Reichardt AD, Cheng G. Non-canonical NF-κB signaling activation and regulation: principles and perspectives. Immunol Rev. 2011;244(1):44–54. doi: 10.1111/j.1600-065X.2011.01059.x. [DOI] [PubMed] [Google Scholar]
- 17.Sun S-C. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21(1):71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai L, Aye Thu C, Liu X-Y, Xi J, Cheung PCF. TAK1, more than just innate immunity. IUBMB Life. 2012;64(10):825–834. doi: 10.1002/iub.1078. [DOI] [PubMed] [Google Scholar]
- 19.Yang D, Tan M, Wang G, Sun Y. The p21-dependent radiosensitization of human breast cancer cells by MLN4924, an investigational inhibitor of NEDD8 activating enzyme. PLoS ONE. 2012;7(3):e34079. doi: 10.1371/journal.pone.0034079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emanuele MJ, Elia AEH, Xu Q, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147(2):459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36(Pt 5):802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 22.Wei D, Li H, Yu J, et al. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res. 2012;72(1):282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32(1):21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6(1):9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, Tan M, Duan H. Antioxid Redox Signal; SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. 2001;3(4):635-650. [DOI] [PubMed] [Google Scholar]
- 26.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 27.Zaharik ML, Nayar T, White R, et al. Genetic profiling of dendritic cells exposed to live- or ultraviolet-irradiated Chlamydia muridarum reveals marked differences in CXC chemokine profiles. Immunology. 2007;120(2):160-172. [DOI] [PMC free article] [PubMed]
- 28.Brownell JE, Sintchak MD, Gavin JM, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37(1):102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 29.Luo Z, Yu G, Lee HW, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72(13):3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 30.Swords RT, Kelly KR, Smith PG, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115(18):3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 31.Grauer O, Wohlleben G, Seubert S, Weishaupt A, Kämpgen E, Gold R. Analysis of maturation states of rat bone marrow-derived dendritic cells using an improved culture technique. Histochem Cell Biol. 2002;117(4):351–362. doi: 10.1007/s00418-002-0384-4. [DOI] [PubMed] [Google Scholar]
- 32.Torri A, Beretta O, Ranghetti A, Granucci F, Ricciardi-Castagnoli P, Foti M. Gene expression profiles identify inflammatory signatures in dendritic cells. PLoS ONE. 2010;5(2):e9404. doi: 10.1371/journal.pone.0009404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barr TA, Brown S, Ryan G, Zhao J, Gray D. TLR-mediated stimulation of APC: Distinct cytokine responses of B cells and dendritic cells. Eur J Immunol. 2007;37(11):3040–3053. doi: 10.1002/eji.200636483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coghill JM, Sarantopoulos S, Moran TP, Murphy WJ, Blazar BR, Serody JS. Effector CD4+ T cells, the cytokines they generate, and GVHD: something old and something new. Blood. 2011;117(12):3268–3276. doi: 10.1182/blood-2010-12-290403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamatos NM, Carubelli I, van de Vlekkert D, et al. LPS-induced cytokine production in human dendritic cells is regulated by sialidase activity. J Leukoc Biol. 2010;88(6):1227–1239. doi: 10.1189/jlb.1209776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herter JR, Fuchs SY. Recognition of substrate and Skp1 by the homologue of slimb (HOS) ubiquitin ligase receptor D role of the F-box. Med Sci Monit. 2002;8(8):BR283–BR288. [PubMed] [Google Scholar]
- 37.Guindi C, Ménard M, Cloutier A, et al. Differential role of NF-κB, ERK1/2 and AP-1 in modulating the immunoregulatory functions of bone marrow-derived dendritic cells from NOD mice. Cell Immunol. 2012;272(2):259–268. doi: 10.1016/j.cellimm.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 38.Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. Transcriptional profiling of the LPS induced NF-kappaB response in macrophages. BMC Immunol. 2007;8:1. doi: 10.1186/1471-2172-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh S. Handbook of Transcription Factor NF-kappaB. Boca Raton, FL: CRC Press; 2007. [Google Scholar]
- 40.Viatour P, Merville M-P, Bours V, Chariot A. Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem Sci. 2005;30(1):43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Milhollen MA, Traore T, Adams-Duffy J, et al. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-kappaB-dependent lymphoma. Blood. 2010;116(9):1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 42.Brunton LL, Lazo JS, Parker KL. Goodman and Gilman’s: The Pharmacological Basis of Therapeutics. 11th ed. New York: McGraw-Hill Professional; 2006. [Google Scholar]
- 43.Coutinho AE, Chapman KE. The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol Cell Endocrinol. 2011;335(1):2–13. doi: 10.1016/j.mce.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Bosscher K, Vanden Berghe W, Haegeman G. The interplay between the glucocorticoid receptor and nuclear factor-kappaB or activator protein-1: molecular mechanisms for gene repression. Endocr Rev. 2003;24(4):488–522. doi: 10.1210/er.2002-0006. [DOI] [PubMed] [Google Scholar]
- 45.Chang F-M, Reyna SM, Granados JC, et al. Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J Biol Chem. 2012 doi: 10.1074/jbc.M112.397703. 287(42):35756-35767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zinser E, Rössner S, Littmann L, Lüftenegger D, Schubert U, Steinkasserer A. Inhibition of the proteasome influences murine and human dendritic cell development in vitro and in vivo. Immunobiology. 2009;214(9-10):843–851. doi: 10.1016/j.imbio.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 47.Gottipati S, Rao NL, Fung-Leung W-P. IRAK1: a critical signaling mediator of innate immunity. Cell Signal. 2008;20(2):269–276. doi: 10.1016/j.cellsig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 49.Pharmaceuticals M. Study of MLN4924 in Adult Patients With Nonhematologic Malignancies. Study of MLN4924 in Adult Patients With Nonhematologic Malignancies.
- 50. MLN4924 for the Treatment of Acute Myelogenous Leukemia, Myelodysplastic Syndrome, and Acute Lymphoblastic Leukemia. 2009.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.