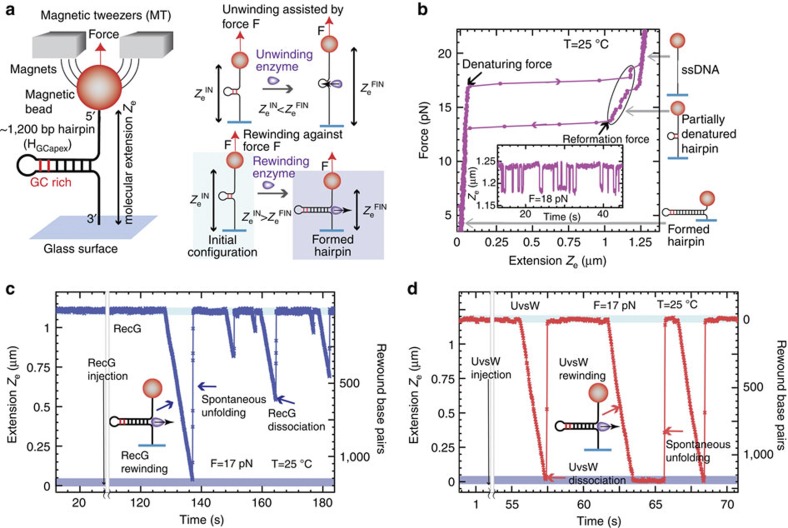
Figure 1. Efficient rewinding of DNA is monitored in real time using magnetic tweezers.
(a) Schematic of the MT experimental setup and progress of the reaction. A DNA hairpin substrate was tethered between a glass surface and a magnetic bead held in a magnetic tweezers. The GC-rich region of the HGCapex hairpin is shown in red. The DNA rewinding or unwinding reaction was followed by monitoring changes in the distance between the bead and the glass surface (Ze). (b) Hairpin configurations can be precisely followed using MT, and typical data in the absence of protein are shown. The force as a function of Ze for the HGCapex hairpin at 25 °C demonstrated stable hairpin folding below 15 pN and mechanical unfolding above 17 pN. Inset shows the Ze(t) at 18 pN where the molecule hops between the fully open (that is, completely denatured) and partially denatured hairpin configurations (c,d). Representative traces of rewinding reactions catalysed by RecG (c) and UvsW (d) enzymes. Reactions were carried out as described in Methods. Rewinding bursts were detected as decreases in Ze. The molecular extensions corresponding to the initial partially denatured hairpin and the final fully formed hairpin are highlighted in light and dark blue, respectively.