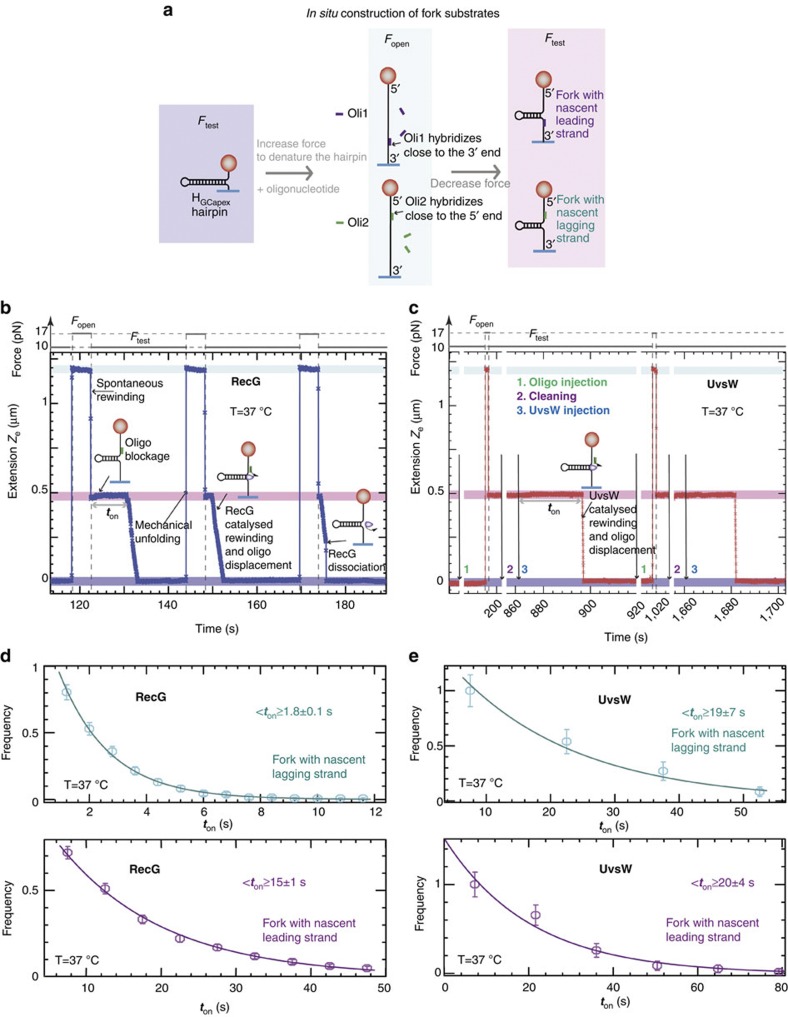
Figure 4. Substrate requirements for RecG- and UvsW-catalysed fork regression are distinct.
(a) Schematics of the forked-DNA substrate construction with nascent lagging or leading strand from the HGCapex hairpin. (b,c) Experimental traces showing the cyclic fork regression assay for RecG (b) and UvsW (c) proteins at 37 °C. The molecular extensions corresponding to the initially formed hairpin, the totally denatured substrate and the partially denatured hairpin are highlighted in dark blue, light blue and pink, respectively. For RecG, a single injection of oligonucleotide and enzyme is sufficient, whereas for UvsW measurements three separate injections are needed: first oligonucleotide injection, second buffer injection and third UvsW injection. (d,e) The distributions of enzyme-binding times for RecG (d), (number n of experimental traces analysed from 427 to 566 depending on the conditions) and UvsW (e), (n from 55 to 86 depending on the conditions) and different fork geometries. Error bars are inversely proportional to the square root of the number of points for each bin.