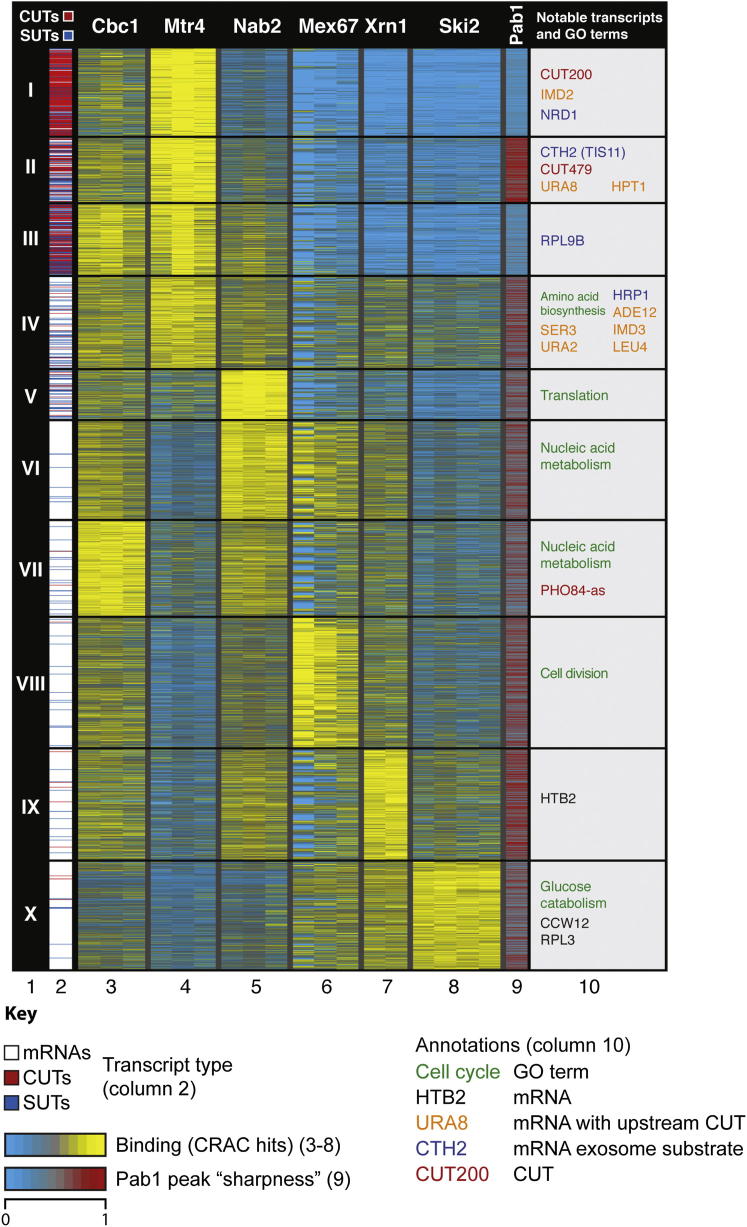
Figure 2.
RNP Composition Reflects Transcript Function and Reveals Distinct RNA Classes
Transcripts arranged by k-medians clustering (n = 4960; k = 10, column 1) based upon their binding to maturation and turnover factors. Column 2: location of CUTs (red), SUTs (blue) and mRNAs (white). Columns 3–8: relative number of hits (per million hits in Pol II transcribed RNAs) in Cbc1, Mtr4, Nab2, Mex67, Xrn1, and Ski2 data sets for each transcript. Column 9: Pab1 peak score, reflecting the specificity of Pab1 interaction. Column 10: significantly overrepresented GO terms (green) and individual transcripts referred to in the text (color coding is indicated in the key). See also Figure S2 and Tables S2, S3, S4.