Figure 4.
Dual Roles of Hrp1 and Nab2 in mRNA Cleavage/Polyadenylation and in CUT Surveillance
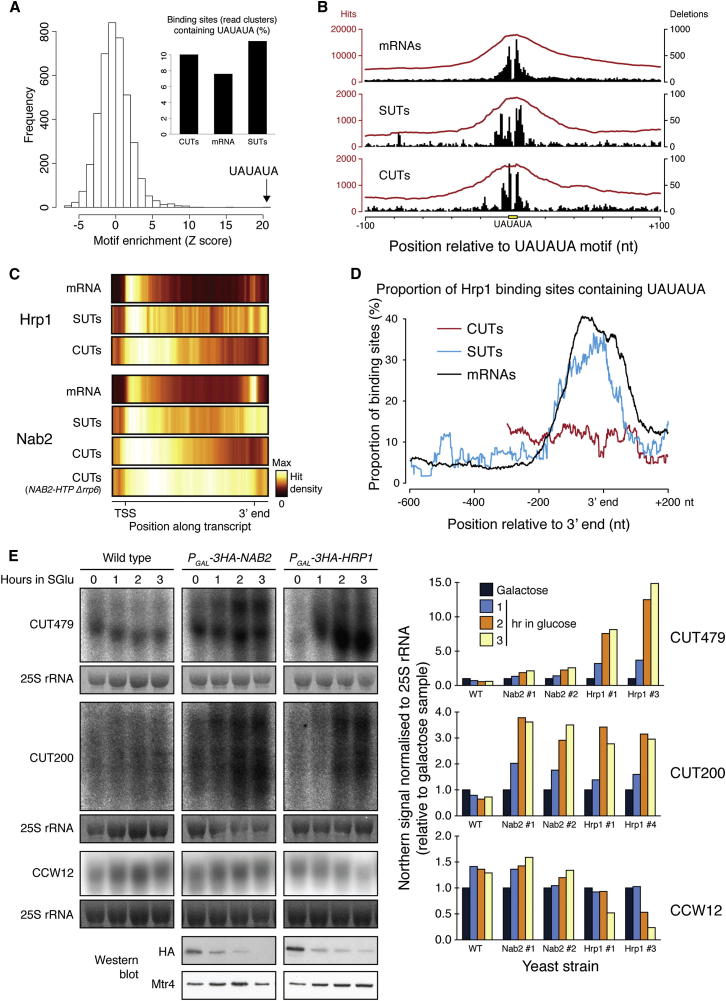
(A) Enrichment scores for 6 nt motifs in Hrp1-bound RNA fragments. Inset: proportion of Hrp1-binding sites containing the UAUAUA motif in mRNAs, CUTs and SUTs.
(B) Distribution of Hrp1 hits (black) and deletions (red) around UAUAUA motifs in mRNAs, CUTs, and SUTs.
(C) Average distribution of binding sites for Hrp1 (top) and Nab2 (bottom) across mRNAs, SUTs, and CUTs in the wild-type background. For Nab2 binding to CUTs an rrp6Δ strain is also shown.
(D) Frequency of UAUAUA motifs in Hrp1-binding sites near the 3′ end of mRNAs, CUTs, and SUTs (Xu et al., 2009).
(E) Northern analysis of CUT479, CUT200, and CCW12 abundance in wild-type, PGAL-NAB2 and PGAL-HRP1 strains after glucose-dependent repression. Northern signals were quantified for replicate experiments (right). Bottom: western analysis of HA-Hrp1, HA-Nab2, and Mtr4 abundance.