Figure S2.
Functions of RNA-Binding Proteins in the Biogenesis and Turnover of mRNAs and lncRNAs, Related to Figure 2
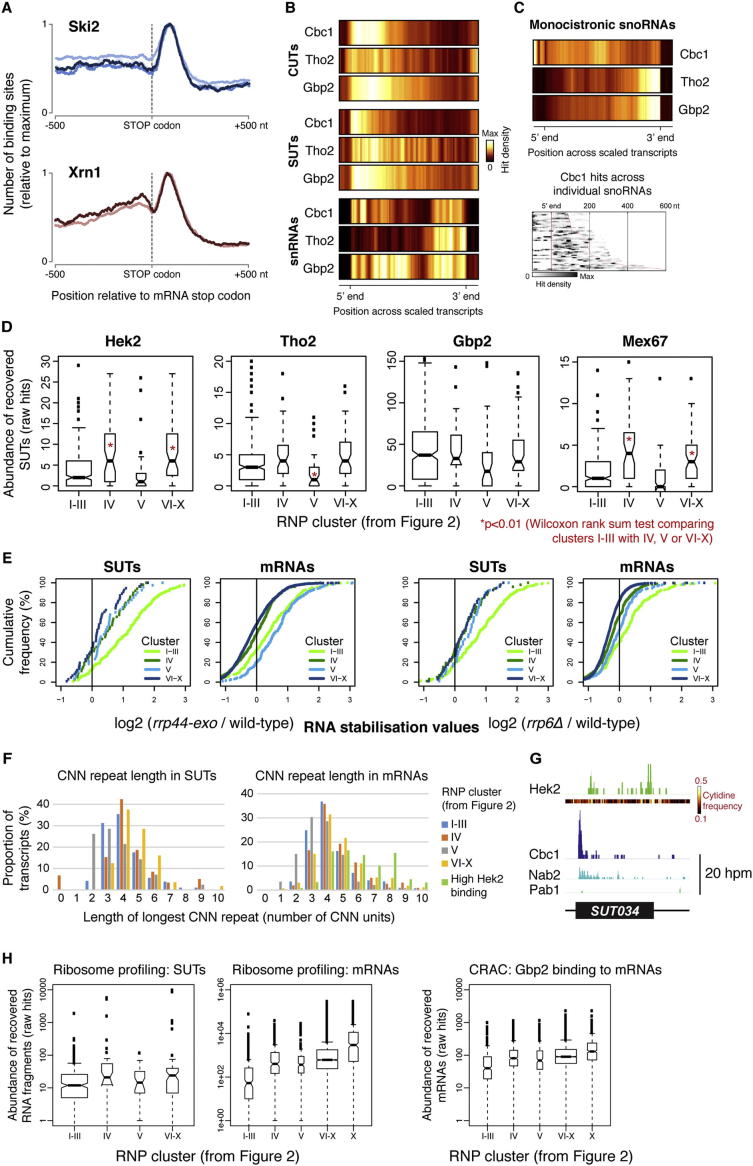
(A) Distribution of Ski2- and Xrn1-binding sites around all mRNA stop codons. Overlapping reads are flattened into single contigs to avoid bias toward abundant transcripts.
(B) Average binding distribution of the cap-binding complex subunit Cbc1 and TREX components Gbp2 and Tho2 across CUTs, SUTs and snRNAs. Average hit densities were calculated for 120 bins spanning the 500 most abundantly bound CUTs or SUTs, and all snRNAs (including 2x10 bins for 100 nt flanking regions).
(C) Top: average binding distribution of Cbc1, Gbp2 and Tho2 across all monocistronic snoRNAs. Average hit densities were calculated for 120 bins (including 2x10 bins for 100 nt flanking regions). Bottom: Cbc1 hits across individual snoRNAs (distance in nt from the 5′ end of the mature snoRNA).
(D) Box and whisker plot of the number of hits for each SUT recovered bound to Hek2, Tho2, Gbp2 and Mex67. Transcripts are grouped by RNP profiles (see Figure 2). Asterisks indicate where median values are significantly different from the cluster I-III median (Wilcoxon rank sum test, p < 0.01).
(E) Cumulative distribution of log2 stabilization ratios for SUTs and mRNAs in the indicated surveillance mutants (Gudipati et al., 2012). Transcripts are grouped by RNP profiles (Figure 2).
(F) Histogram showing the prevalence of SUTs (left) or mRNAs (right) with different length (CNN)n motifs. For each transcript, the longest CNN repeat was recorded, and the proportion of transcripts with each CNN repeat length plotted. Transcripts were divided into the clusters defined in Figure 2. For mRNAs, an additional category is included (green), comprising mRNAs with particularly high binding to Hek2 (determined by clustering analysis as per Figure 2, but with Hek2 included).
(G) Distribution of Hek2, Cbc1, Nab2 and Pab1 CRAC hits across SUT034. Cytidine frequency within the SUT sequence is indicated.
(H) Box and whisker plot of the total number of hits for each SUT or mRNA recovered in ribosome profiling experiments (Brar et al., 2012) (GEO sample GSM843748). Transcripts are divided into the clusters defined in Figure 2. Although cluster X mRNAs are included in the cluster VI-X category, we also show them separately as they appear to be particularly highly translated. For comparison, the number of Gbp2 CRAC hits for each mRNA is shown (right). The upper and lower edges of the boxes are the upper and lower quartiles, respectively. The whiskers extend from these edges to the most extreme value within 1.5 times the length of the box.