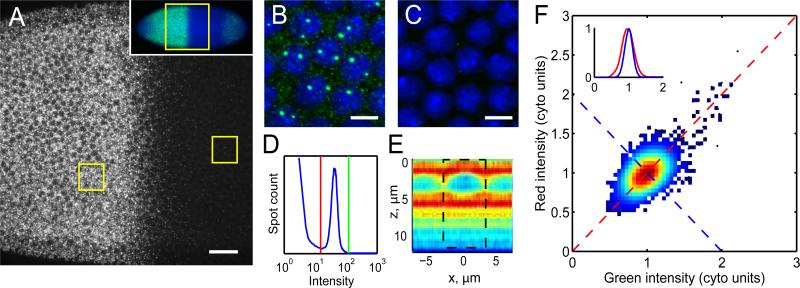
Figure 1. Counting of absolute transcript number in Drosophila embryos.
A: Confocal section through the nuclear layer of a WT embryo during interphase 13 labeled with 114 fluorescent oligonucleotide probes against hunchback, oriented anterior to the left. Scale bar: 25 μm. Inset: Low magnification image identifying the region shown in A. B,C: Magnified views of anterior (B) and posterior (C) boxed regions in (A). Scale bars: 5 μm. D: Particle intensity histogram showing thresholds separating transcripts from noise (red line) and from the long tail of bright transcription sites (green line). E: hb transcript distribution in axial cross-section through a nucleus centered at x=0. z=0 represents apical surface. Color indicates mean particle density in relative units (red=high, blue=low). Dashed box: cylindrical summation volume. F: Intensity scatter plot in two channels using probes of alternating colors. Data point density given by color. Inset: Cross-sections of scatter plot in (F) along the correlated (red) and anti-correlated direction (blue) shows Gaussian distributions with σ=20% (red) and σ=12% (blue) after normalization to mean cytoplasmic particle intensity (1 “cyto unit”). See also Figures S1 and S2.