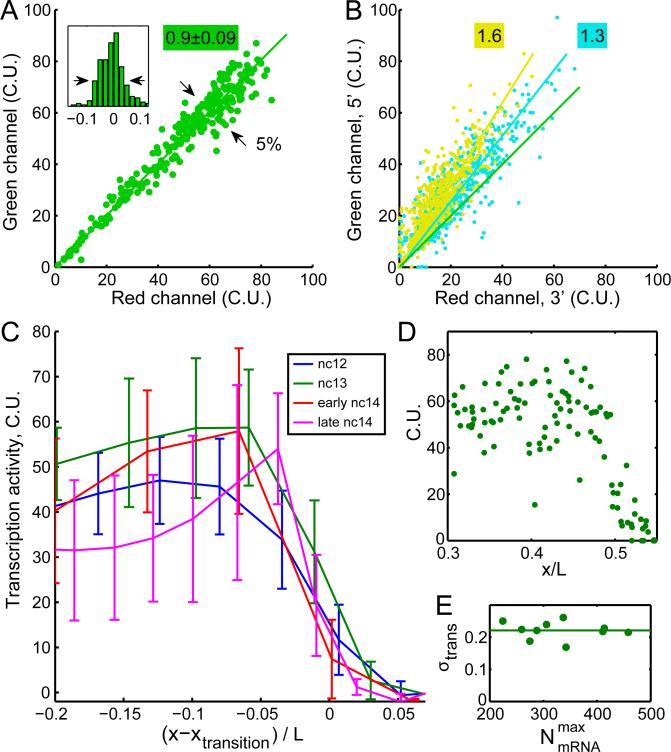
Figure 3. Variability of transcriptional activity at nascent transcription sites.
A: Scatter plot of total nascent hb mRNA per nucleus using probes of alternating colors for an embryo in nuclear cycle 13 after normalization to the mean cytoplasmic particle intensity (C.U.). Intensities follow a direct proportionality relation with slope 0.90±0.09 (n=5 embryos). Inset: root mean square normalized deviation from linear fit; scatter = 5% (arrows). B: Two-color scatter plot of nascent mRNA content in which probes bearing the same fluorophore are clustered on the 5’ (green channel) and 3’ (red channel) portions of the transcript. Cyan: measurements using 57 green and 57 red-labeled probes; observed slope: 1.3. Yellow: results with 78 green and 36 red-labeled probes; observed slope: 1.6. Green line in A is plotted for comparison. C: Transcriptional activity per nucleus as a function of position along the AP axis for four embryos in nuclear cycle 12 (blue), 13 (green), 14 early (red), and 14 late (magenta) in binned averages of 10, 20, 40 and 40 nuclei, respectively. Error bars: SDs within bins. Position is shown as distance from inflection point xtransition. D: Transcriptional activity per nucleus as a function of absolute AP position for the embryo in interphase 13 in C. E: Transcription noise for 10 embryos in nuclear cycle 13 plotted as fractional SD across nuclei as a function of cytoplasmic hb counts within the spatial domain of highest accumulation. Transcription activity noise remains constant throughout interphase at 22±3%. See also Figure S4.