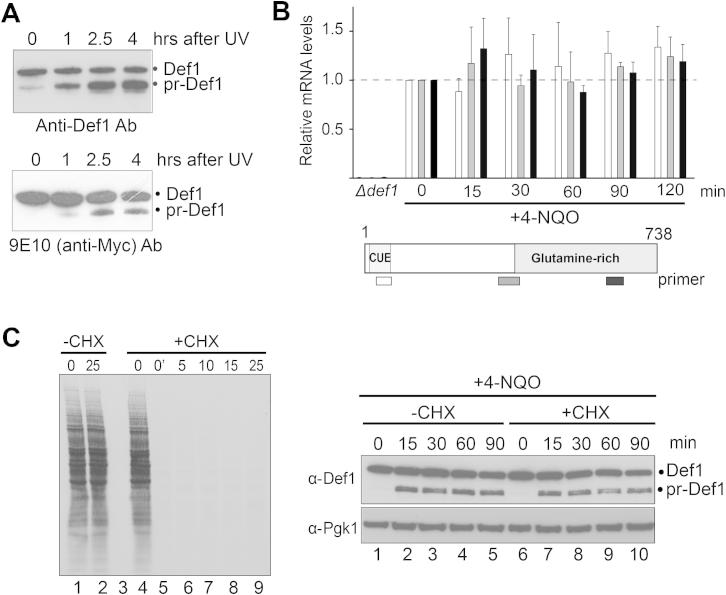
Figure S1.
pr-Def1 Is Not Formed from Alternate Transcription or Translation, Related to Figure 1
(A) pr-Def1 may in some cases appear to be generated without equivalent disappearance of full-length Def1. This is due to the anti-Def1 antibody exhibiting an unusual and pronounced preference for pr-Def1 over the full-length protein: only a small fraction of Def1 is actually processed, making it difficult to appreciate its depletion. To illustrate this point, the same samples from cells expressing N-terminally Myc-tagged Def1 were subjected to western blotting and probed with either polyclonal anti-Def1 antibodies (upper panel), or anti-Myc antibodies (lower panel). Note the strong preference for pr-Def1 exhibited by the anti-Def1 antibody (upper), which gives the false impression that pr-Def1 may not have been generated from full-length protein, which is only slightly depleted at the same time-points. However, weak, concomitant depletion of full-length Def1 can be appreciated by the use of anti-Myc antibody (lower). Because introducing N-terminal tags in DEF1 in all of the many strains used in this study would be impractical, Def1 processing was detected with the polyclonal Def1 antibody unless otherwise stated.
(B) Total RNA was isolated, reverse transcribed, and relative DEF1 mRNA levels were measured by qPCR, in the relevant areas of the transcript, before and after DNA damage. Quantification was normalized to β-actin mRNA levels in each sample, then against untreated conditions (time0). Primers hybridized to the start (white), middle (gray), and end (black) of the DEF1 mRNA. Error bars correspond to the standard deviation across three biological replicates. None of the mRNA areas, in particular the region encoding Def1’s C terminus, are significantly up- or downregulated in response to DNA damage and Def1 processing.
(C) Def1 processing occurs posttranslationally. (left) Autoradiogram of SDS-PAGE gel, showing cellular proteins, radioactively labeled in vivo. Protein synthesis was inhibited by the addition of 25 μg/ml cycloheximide (CHX) to midlog cells at time0, and new protein synthesis measured by incorporation of radioactive label in proteins (10 min S35-methionine pulses initiated at the times indicated after cycloheximide inhibition). Time-point 0’ was transferred to grow in radioactive methionine immediately after the addition of cycloheximide. Note that cycloheximide completely, and immediately, inhibits new protein synthesis. (Right) Western blot showing that Def1 processing occurs even in the presence of effective cycloheximide inhibition of new protein synthesis (compare lanes 7-10 with 2-5). Cycloheximide was added 5 minutes prior to the addition of 4-NQO.