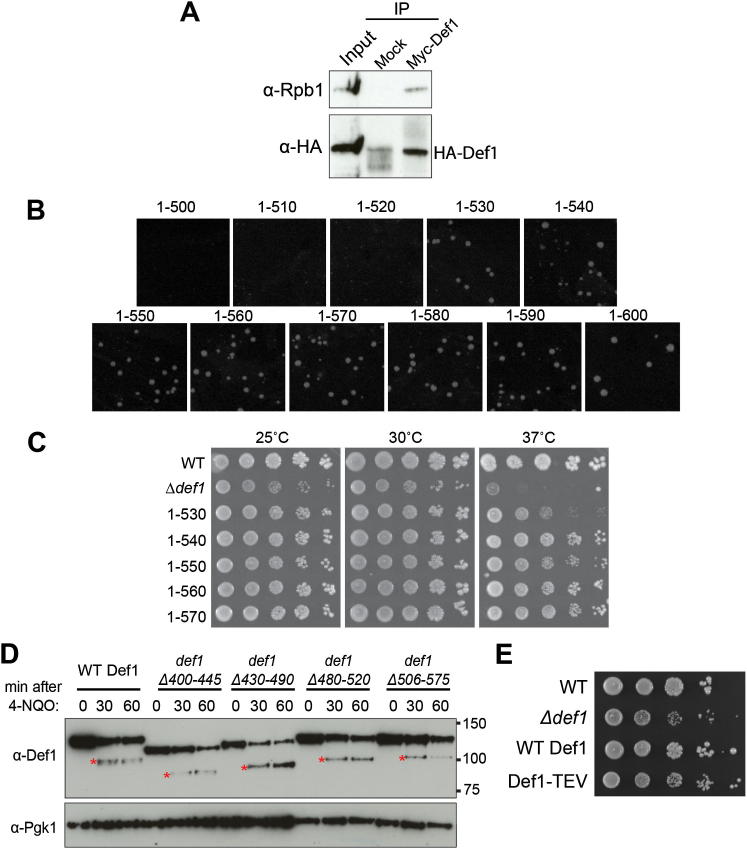
Figure S2.
Mapping the Site of Def1 Processing, Related to Figure 2
(A) Western blot showing Def1-HA and Rpb1 retained on immobilized Myc-Def1 or mock beads after immuno-precipitation in extracts derived from cells expressing both Myc-Def1 and Def1-HA.
(B) Yeast minimal media plates showing viability of transformed cells. Cells were transformed with equal amounts of PCR product for genomic truncation of Def1 at 10 amino acid intervals between 500-600 amino acids, as indicated. This simple procedure for comparing viability after transformation of similar amounts of slightly different DNA constructs is highly reliable and reproducible when performed on the same transformation-competent, yeast culture.
(C) Dilution series of yeast cells of the genotype indicated on the left, grown at 25°C, 30°C and 37°C.
(D) Def1 internal deletions result in the processing site shifting to another position. Western blots of extracts from WT Def1 and the indicated Def1 internal deletion strains, before and after treatment with 4-NQO for 30 and 60 min. The exact site of processing is not fixed, as the size of pr-Def1 changes (red asterisks). The mobility of the full-length Def1 proteins vary more than would be expected based on the difference in amino acids number between them. This is because glutamine-rich regions - deleted in some, but not others - contribute disproportionally to SDS-PAGE mobility.
(E) Def1-TEV does not exhibit growth defect. Δdef1 cells are slow-growing (second line of cell dilution), but not when expressing WT, or Def1-TEV (lower lines). Def1-TEV can also still be normally processed in response to transcription stress (data not shown).