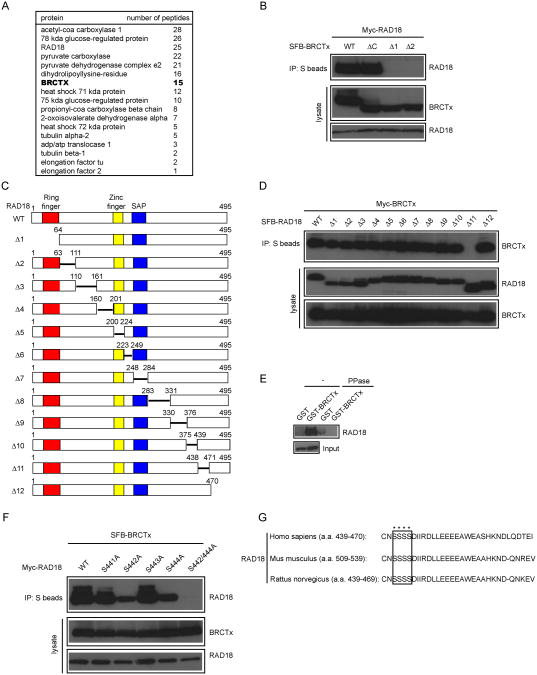
Fig.2. BRCTx interacts with RAD18.
(A) 293T cells stably expressing SFB-tagged-BRCTx were used for tandem affinity purification of protein complexes specifically from chromatin fractions. Tables are summaries of proteins identified by mass spectrometry analysis. Letters in bold indicate the bait proteins. (B and D) Mapping of the corresponding regions required for the BRCTx-RAD18 interaction. Immunoprecipitation reactions were performed using S-protein beads and then subjected to western blot analyses using antibodies as indicated. (C) Schematic representation of wild-type and deletion mutants of RAD18 used in this study. (E) Phosphorylation-dependent interaction between BRCTx and RAD18. GST or GST–BRCTx was incubated with cell lysates containing exogenously expressed Flag-tagged wild-type RAD18, with or without phosphatase (PPase). Bound RAD18 was analyzed by anti-Flag immunoblotting. Lower gel shows amounts of RAD18 protein used in these experiments. (F) Both the Ser442 and Ser444 residues of RAD18 are required for its binding to BRCTx. 293T cells were transfected with plasmids encoding SFB-tagged BRCTx together with plasmids encoding Myc-tagged wild-type or serine point mutants of RAD18. Cells were collected 24 h after transfection. Coimmunoprecipitation reactions were performed using S-protein beads, and cells were then subjected to immunoblotting using the antibodies as indicated. (G) Alignment of the BRCTx binding region of RAD18 from different species.