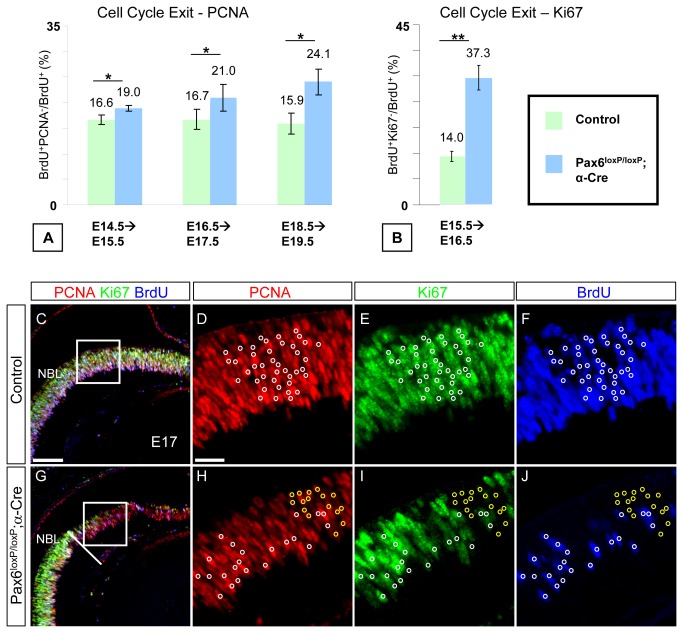
Figure 2. Neurogenic Pax6- RPCs in the Pax6 loxP/loxP ;α-Cre retina display increased cell-cycle exit and sustained expression of cell-cycle factors.
A single pulse of BrdU was administered at E14.5, E15.5, E16.5 and E18.5, 24 h prior to sacrifice as indicated (A, B). Pax6 expression was detected on adjacent sections to identify the recombination area in the Pax6loxP/loxP;α-Cre OC. (A) The percentage of BrdU + PCNA-/BrdU+ Total was determined for the Pax6loxP/loxP control (green bars) and Pax6 loxP/loxP ;α-Cre (blue bars) distal retina. Pax6 - RPCs show increased cell-cycle exit at all stages tested (19% (SD=0.6%), 21% (SD=2.7%) and 24.1% (SD=2.6%) compared to 16.6% (SD=0.93%), 16.7% (SD=2%) and 15.9% (SD=2%) in control at E14.5, E16.5 and E18.5, respectively; p≤0.05, n≥3). (B) The number of BrdU+ and BrdU + Ki67- cells was quantified and the ratio BrdU + Ki67-/BrdU+ Total was used to compare cell-cycle exit rate in control (green bar) and Pax6 loxP/loxP ;α-Cre (blue bar) distal retina at E15.5. Pax6 - RPCs show increased cell-cycle exit (37.3% (SD=3.6%) in Pax6 - compared to 14% (SD=1.5%) in control; p<0.01, n=6 for both genotypes). Triple immunofluorescence for PCNA, Ki67 and BrdU (red, green and blue, respectively, in C–J) in control (C–F) and Pax6 loxP/loxP ;α-Cre (G–J) distal retina showing mitotic PCNA + Ki67 + BrdU+ in both control and mutant (white circles in D–F,H–J) and abnormal PCNA + Ki67-BrdU- cells detected only in Pax6 loxP/loxP ;α-Cre (yellow circles in H–J) OC. Scale bars in C,D are 100 and 25 µm, respectively.