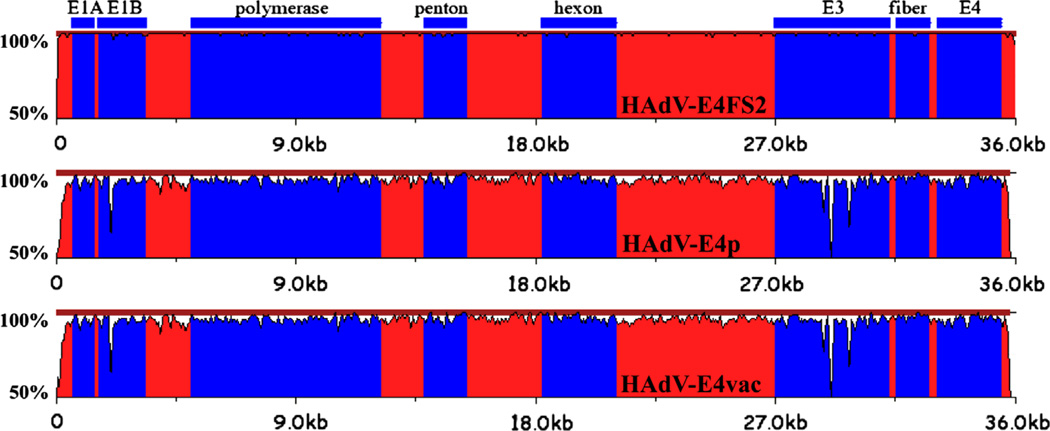
Fig. 1.
Pairwise genome comparative analysis. zPicture (http://zpicture.dcode.org/) utilizes a local alignment algorithm, BlastZ, to display the regions of nucleotide sequence similarity between HAdV-E4FS1 and query genomes: HAdV-E4FS2 (top); HAdVE4p (middle); and HAdV-E4vac (bottom). Of note are the divergences at the ends of the genomes, corresponding to the semi-conserved inverted terminal repeat (ITR) sequences that contain critical functions, including DNA replication. The genome sequences are arrayed on the horizontal and the percent identity of genome pairs from 50% to 100% is noted along the y-axis. The colors are arbitrary and are used to provide contrast: blue regions highlight select genes or genome regions (E3), noted above the alignments. Red regions include both coding and noncoding sequences.