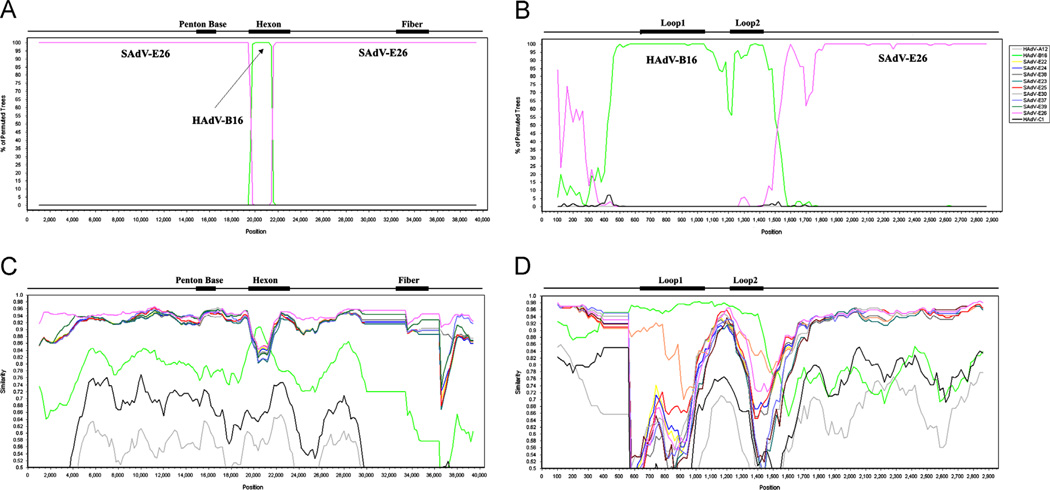
Fig. 3.
Nucleotide sequence recombination analysis of the whole genome and the hexon gene of HAdV-E4FSl. The genome of HAdV-E4FSl (AY599837) was analyzed for sequence recombination events with HAdV and SAdV genomes using the software Simplot, which is available on-line (http://sray.med.som.jhmi.edu/SCRoftware/simplot/). Bootscan analysis of the genome (A) and hexon (B) shows a lateral transfer of a portion of the hexon gene from HAdV-B 16. These are supported by the Simplot analysis of the genome (C) and hexon (D). Unlike the Simplot panels (C and D), only the SAdV-E26 genome, rather than the entire set of SAdV-E22 through -E26 sequences, is presented in the Bootscan panels (A and B), as similar genomes compete out the signal. For Bootscan, removing four of the five genomes and repeating the analysis with each individually, gives a clearer representation, reflecting the Simplot result for the highest similarity match. MAFFT software was used to align the sequences prior to recombination analysis (http://mafft.cbrc.jp/alignment/server/). Default parameter settings for the Simplot software were used for analyzing the hexon sequences: window size (200 nucleotides [nt]), step size (20 nt), replicates used (n=100), gap stripping (on), distance model (Kimura) and tree model (neighbor-joining). Similarly, whole genomes were analyzed, beginning with an initial alignment using MAFFT and following with recombination analysis using Simplot and Bootscan. Only the window size and step size were altered for the genome analysis (1000 and 200, respectively), with the rest of the default parameters unchanged. Genome nucleotide positions are noted along the x-axis, and the percentages of permutated trees that supported grouping are marked along the y-axis. For reference, select genome and gene-specific landmarks are noted above each graph. Colors: pink, SAdV-E26 and green, HAdV-B16.