FIGURE 4.
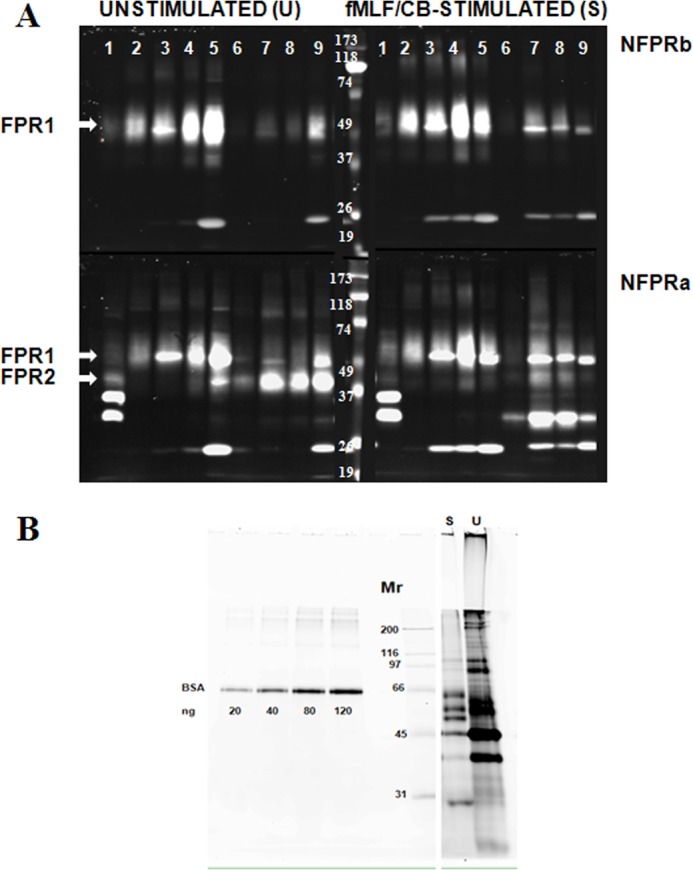
Partial purification of FPR1 and FPR2 from DDM lysates of unstimulated and fMLF/cytochalasin B-stimulated neutrophils by serial immunoaffinity capture using NFPRb- and NFPRa-Sepharose. A, serial affinity purification of FPR1 and FPR2. U and S neutrophils prepared as described under “Experimental Procedures” were lysed in DDM-containing lysis buffer at 2 × 108 cells/ml, precleared of debris antibody binding components first by centrifugation at 17,000 × g and then by passage over an isotype-matched mAb CS9 affinity matrix. The extracts (lane 1) were then serially exposed to NFPRb-Sepharose followed by exposure of the flow-through to an NFPRa-Sepharose matrix. The NFPRb matrix (lanes 2–5) and the NFPRa matrix (lanes 6–9) were eluted with first epitope mimetic peptide A&B buffer (lane 2) and then a low pH buffer (lane 3) and 1% SDS-containing buffer (lane 4). The NFPRa matrix was similarly eluted with mimetic peptide C&D buffer (lane 6), followed by a low pH buffer (lane 7) and 1% SDS-containing buffer (lane 8). The washed matrix beads were then put into SDS-PAGE sample buffer, heated, centrifuged, and run in lanes 5 (NFPRb-beads) and 9 (NFPRa-beads). Mr markers are shown in the center panel of the bottom row. Only the peak fractions are shown. B, protein composition of NFPRb peptide eluates. 10-μl samples of each NFPRb eluate U and fMLF/CB were labeled with 600 pmol of fluorescence protein Z dye Blue 2 (see “Experimental Procedures”) as described previously using similarly labeled bovine serum albumin as a standard. Major bands were tryptically digested and identified by Agilent ESI quadrupole-TOF mass analysis in Table 1.
