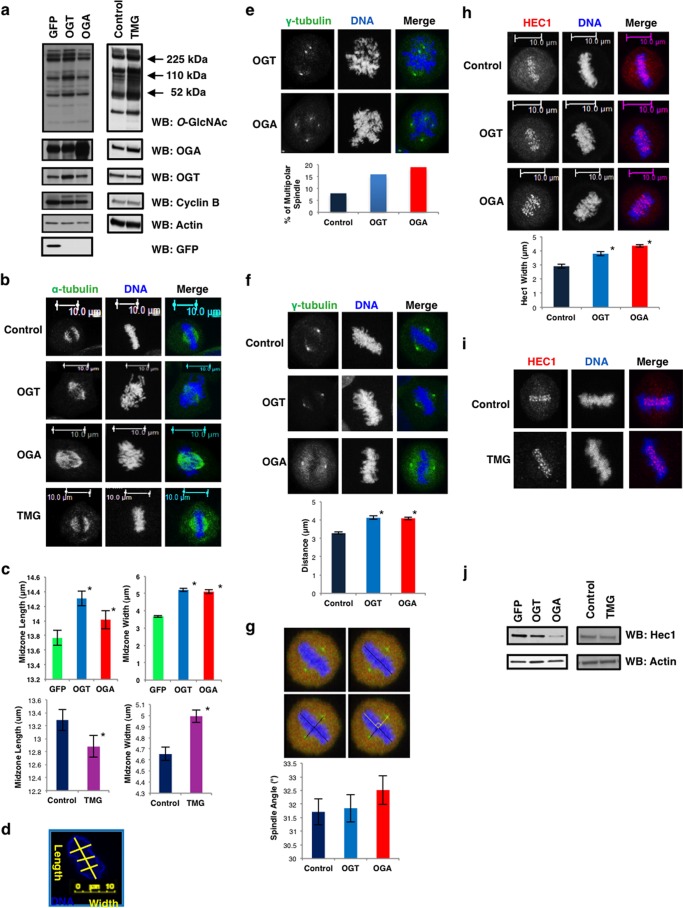
FIGURE 1.
OGT or OGA gain of function disrupts spindle architecture. a, GFP, OGT, or OGA gain of function or OGA inhibitor Thiamet-G-treated HeLa cells were synchronized to M phase. Western blots were performed for O-GlcNAc, OGA, OGT, GFP, cyclin B, and actin. b–d, DNA (blue) and α-tubulin (green) confocal imaging of M phase gain of function OGT/OGA cells, or cells treated with Thiamet-G. Midzone width and length (mean ± S.E., replicate number (n): nGFP = 195, nOGT = 193, nOGA = 198, nNT = 195, nTMG = 167, *, p < 0.005 between GFP/NT versus OGT/OGA or TMG) were quantified using ImageJ software as follows: first, the condensed chromatin area color was inverted in order to delineate the edge of the chromatin. Next, the chromatin was measured lengthwise, and the width was measured three times from different sections and averaged. Yellow lines are a representative measurement. e, multipolar spindles were quantified from confocal images of DNA (blue) and γ-tubulin (green) M phase synchronized gain of function OGT/OGA cells. f, DNA (blue) and γ-tubulin (green) were confocal imaged at M phase in OGT/OGA gain of function cells. Distance between each centrosome and the midzone was quantified (mean ± S.E., replicate number (n): nControl = 93, nOGT = 64, nOGA = 76, *, p < 0.005 between Control versus OGT/OGA). g, spindle angle measurement schematic in OGT/OGA gain of function cells. The perpendicular black lines originating from the centrosomes and the center of the spindle midzone were used to calculate spindle angle shown in yellow and plotted as a histogram. h, DNA (blue) and HEC1 (red) were confocal imaged at M phase in OGT/OGA gain of function cells. Average width of HEC1 staining was quantified using ImageJ (mean ± S.E., replicate number (n): nControl = 21, nOGT = 47, nOGA = 50, *, p < 0.005 between Control versus OGT/OGA). i, DNA (blue), α-tubulin (green), and HEC1 (red) were confocal imaged at M phase in TMG-treated cells. j, Western blot of HEC1 and actin in OGT/OGA gain of function and TMG-treated cells.