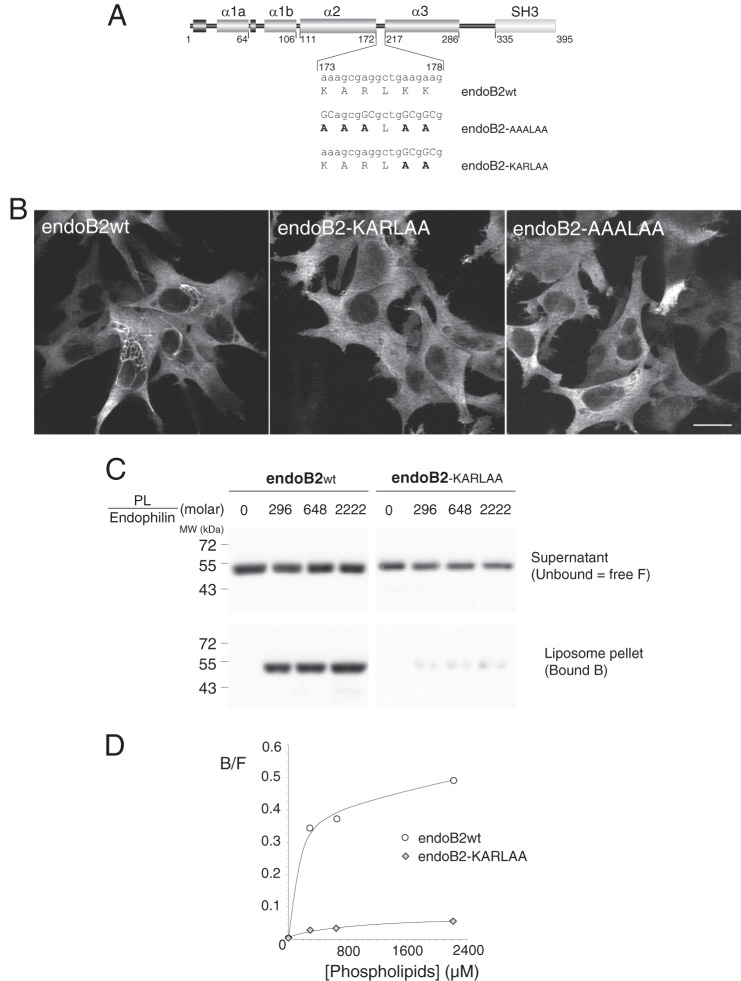
FIGURE 6.
Mutagenesis of critical residues in the BAR sequence involved in membrane binding abrogate the assembly of endoB2 into perinuclear filamentous structures. A, rod structure of endoB2 showing the domain organization and the two mutations used in this study of the 173–178 amino acid loop (KARLKK) between α-helices 2 and 3. Based on the crystal structure of human endophilin BAR domain (Protein Data Bank code 1X03 A), this sequence is located at both tips of the dimer. B, HeLa cells were transfected to express, as indicated, wild type endoB2 or the mutants fused to a C-terminal Myc tag. Images show z-sections obtained from cells processed for immunofluorescence analysis using anti-Myc antibodies. For both mutants, not a single cell on the entire coverslips showed the typical formation of perinuclear filamentous structures as in the case of wild type endoB2. Scale bar, 20 μm. C, membrane binding of wild type (wt) and doubly mutated (KARLAA) species was examined using phospholipid to endophilin molar ratio ranging from 0 (input) to 2222 as indicated. 11.1% of the supernatant (unbound) and 33% of the pellet (bound) (see “Experimental Procedures”) were immunoblotted using separate polyacrylamide gels and anti-Bif antibody. No precipitation of endophilins was observed in the absence of membranes. D, chemiluminescence signals obtained from the blot shown in C and representative of two experiments. Plotted is the bound (B) to free (F, unbound) ratio as a function of the phospholipid to endophilin ratio.