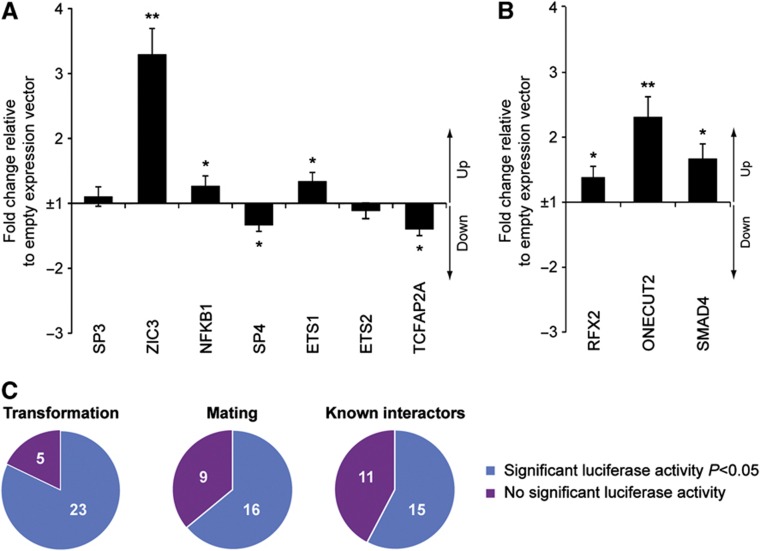
Figure 3.
Luciferase validation of Y1H interactions. Luciferase reporter-based validation with either the Mmp9 promoter (A) or the Mcts2-Id1 enhancer element (B), respectively. The results for the other DNA baits can be found in Supplementary Figure S10. The fold change of the normalized firefly to Renilla ratio compared to the normalized firefly to Renilla ratio of the negative control (empty expression vector) is plotted. Consensus interactions were analyzed as well as those reproducibly found by either mating or transformation. Interactors reported in the literature (Supplementary Table S2) and not found in the Y1H screens were also tested. The error bars represent the standard error of six independent biological experiments. **P<0.01 and 0.01<*P<0.05 compared with the negative control. (C) Proportion of reproducibly detected transformation-, or mating-based interactions that scored positive in the luciferase reporter assay. As a comparison, the proportion of known interactions (i.e., positive controls) that altered luciferase activity is also plotted. Source data for this figure is available on the online supplementary information page.