Figure 7.
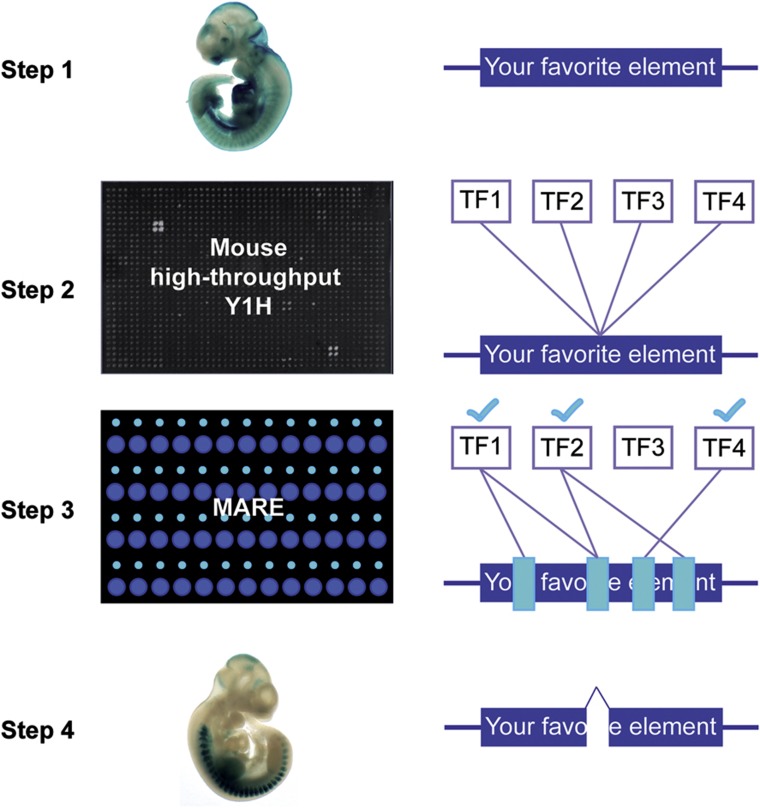
Schematic overview of the pipeline employed to deorphanize mammalian gene regulatory elements. The regulatory element of interest is first cloned (Step 1), and then integrated into yeast to enable high-throughput Y1H screens leading to the identification of putatively interacting TFs (Step 2). In Step 3, MARE analysis is performed to both validate (reflected by light blue check marks) and map the detected TF–DNA interactions (indicated by light blue boxes) within the respective regulatory element. Finally, small binding regions of interest can be deleted to examine the relevance of these DNA segments in mediating the in vivo activity of the regulatory element (Step 4).