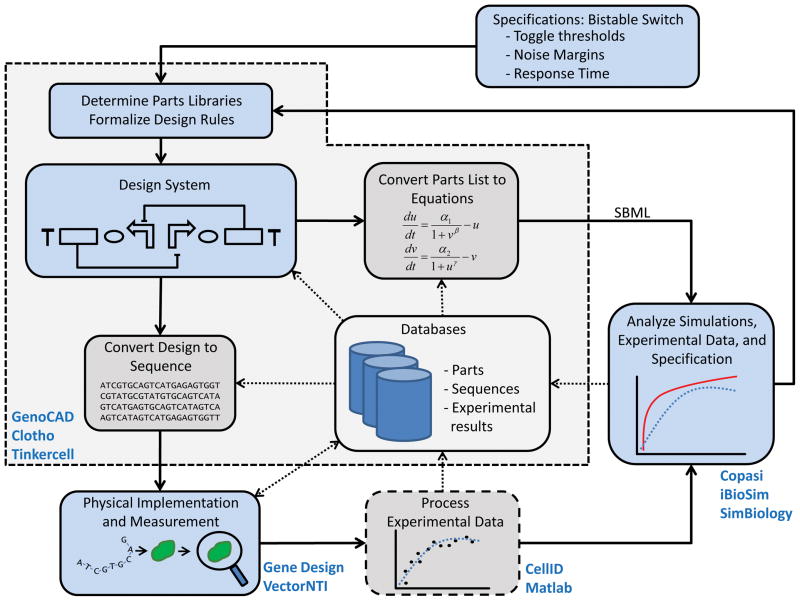
Figure 1.
GDA design flow. Synthetic biology projects typically rely on iterative workflows composed of different tasks. Emerging GDA tool chains rely on numerous software applications that support different phases of the project workflow. The development of a genetic switch [72] will start by expressing the design objective as a list of quantitative requirements: input toggle thresholds, noise margins, switching response time, etc. Once the objective is specified, it is possible to develop a list of genetic parts useable for the project. The choice of biological parts will involve factors such as use of the parts in prior projects, quality of the data characterizing the parts function, or intellectual property considerations. The formalization of design rules often takes place in parallel to the parts library development. Design rules may express rules such as whether it is acceptable to have polycistronic expression cassettes or if the design should be split between different plasmids. Only after parts have been selected and a strategy has been agreed upon is it possible to start designing constructs. In the fabrication phase, the construct is assembled usually by combining de novo gene synthesis and cloning of existing DNA sequences. Users use molecular biology software suites to facilitate assembly or order the sequence from a gene synthesis company. Experimentalists insert the synthetic DNA molecule into the host of choice and collect phenotypic data. Experimental data is then processed, for example by reducing microscopy images into time series of quantitative data. Performance is evaluated by considering simulations, experimental data, and the original specifications. At nearly every stage, software interacts with databases to reuse leverage past work or to store current work for future use. The shaded area delimited by dashes delineates stages facilitated by synthetic biology CAD software, while other stages are handled by more general purpose software. Text in blue indicates examples of software providing assisting at each stage.