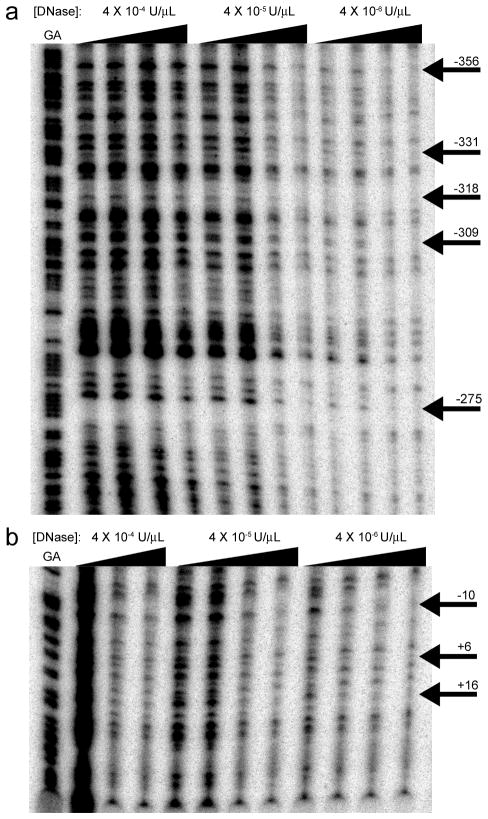
FIGURE 4. DNase protection assay.
Using probes generated from the pGL3-Basic constructs created for the reporter assays in conjunction with various primary cell nuclear B cell extracts (tonsillar, peripheral blood, and splenic B cells, in repetitions of the assay), footprinting regions were identified as those which faded more quickly than surrounding bands as the DNase and protein concentration were adjusted. These footprints are noted by the solid black arrows to the right of the lanes. The far left band is a GA tracker, showing the locations of guanine and adenine bases within the sequence. The concentration of DNase during treatment is noted above, while protein concentration is noted with the wedges of increasing concentration at 0, .013, .065, and .13 mg/mL nuclear extract. A and B, Excerpts from representative exposures of two different probes following DNase digestion, probes spanning regions from −676 to −193 and −243 to +40 relative to the TSS, respectively.