Figure 3.
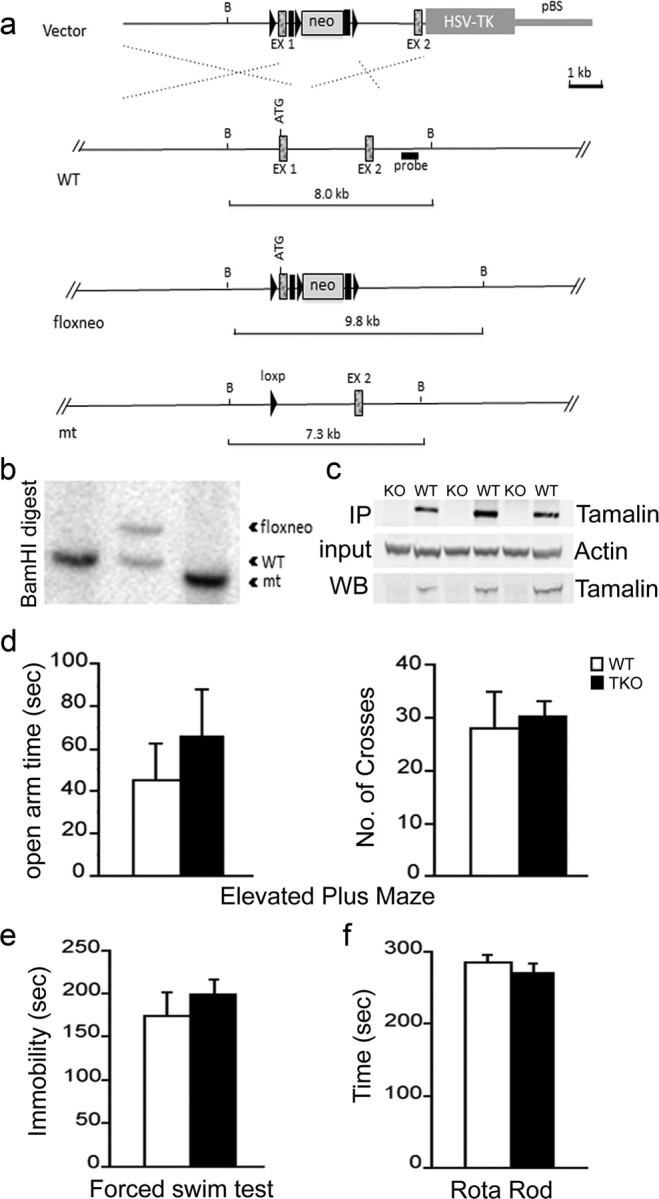
Targeted deletion of tamalin in mouse. Schematic representation of the strategy used to target the tamalin gene. LoxP sites (black triangles) were inserted upstream and downstream of the ATG-containing exon 1 (EX1). The neomycin resistance cassette (neo) is flanked by both loxP and Frt (black rectangles) sites. After the initial screening of the positive ES cell clones, as described in Material and Methods, a probe (black rectangle) downstream of EX1 was used for the identification of the different targeted alleles by detecting a shift from the endogenous BamHI WT band of 8 kb to the rearranged 9.8 kb band resulting from the insertion of the neo cassette by homologous recombination. Cre-induced excise of the tamalin-specific exon and the neo cassette causes a shift of the 9.8 kb band to 7.3 kb. b, Southern blot analysis of genomic DNA from mice with the tamalin alleles indicated in a. Analysis of DNA digested with the BamHI restriction enzyme and probed with the 3′ fragment described in a yields the expected 8 kb band in a WT mouse (lanes 1) and a rearranged 9.8 kb fragment with the neo insertion (floxneo) in an heterozygous mouse and a 7.3 kb band in the mutant (lanes 2 and 3, respectively). c, Western blot analysis of hippocampus dissected from mutant (KO) or WT mice and hybridized with an antiserum directed against tamalin. The top panel is a Western blot analysis of lysates subjected first to immunoprecipitation (IP) with a tamalin-specific antibody and then to the blotting with the same antibody. The bottom panel is the Western blot analysis of straight hippocampus lysates hybridized with the tamalin-specific antibody. The middle blot was hybridized with an anti-β-actin-specific antibody to control for loading. Behavioral analysis of tamalin-deficient mice (d–f). Animals were subjected to the elevated plus maze (d), forced swim (e), and rotarod (f) test. n ≥ 7.
