Fig. 5.
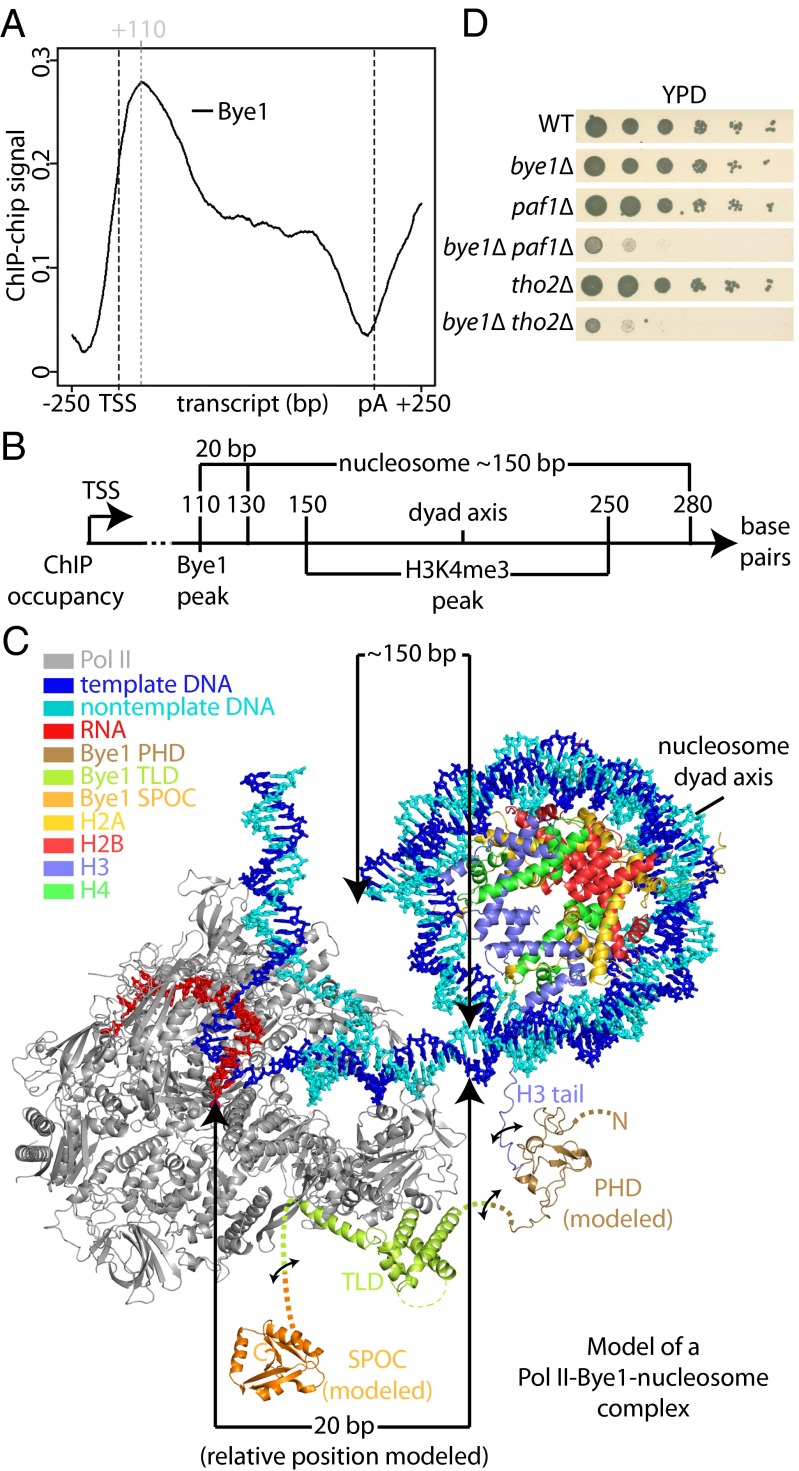
Bye1 associates with active genes in front of the +2 nucleosome. (A) Gene-averaged Bye1 ChIP occupancy profile for the median gene length class (1,238 ± 300 nt, 339 genes). (B) Scheme showing occupancies of Bye1 and H3K4me4 (30) derived from ChIP data and nucleosome position derived from microarray and high-throughput DNA sequencing data (31). (C) Model of a Pol II–nucleosome-Bye1 complex based on crystal structures and ChIP occupancy peak positions. Distances in base pairs (bp) are indicated between the Pol II active center and the nucleosome, as well as for the nucleosomal DNA. The model is based on the structure of the nucleosome core particle (1aoi) (47). Modeling was performed with Coot (48). Bye1 PHD and SPOC domains were modeled using Modeler (49). The PHD domain model is based on structures 3kqi, 1wem, 1wew, 2lv9, and 1wep, which were identified by HHpred (50) to be most similar to Bye1 PHD. Binding of the PHD domain to H3K4me3 was modeled based on structure 2jmj. The SPOC domain model is based on structure 1ow1. (D) Bye1 genetically interacts with Paf1 and Tho2. Serial dilutions of strains bye1∆, paf1∆, bye1∆paf1∆, tho2∆, bye1∆tho2∆, and an isogenic WT control strain were placed on yeast extract peptone dextrose (YPD) plates and incubated at 30 °C for 3 d.