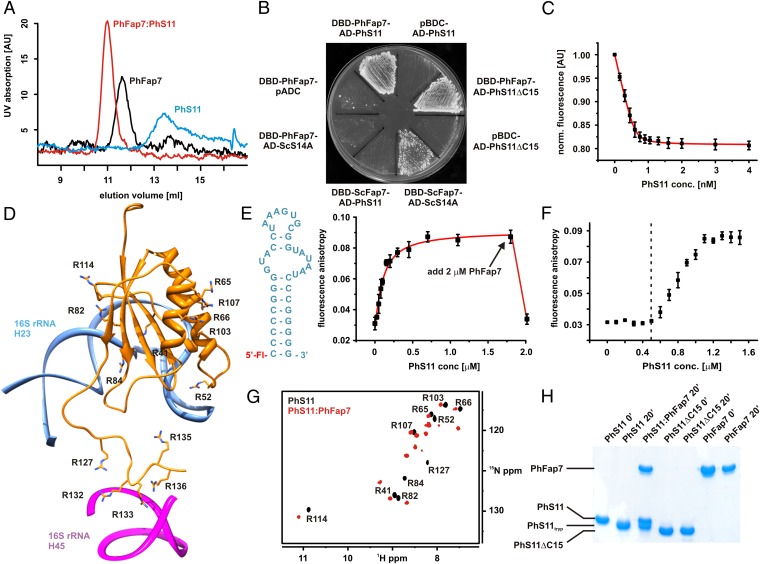
Fig. 1.
PhFap7 binds PhS11 with high affinity and blocks both of its rRNA-binding elements. (A) Gel filtration profiles of PhFap7 (black), PhS11 (blue), and a 1:1 PhFap7/PhS11 (red) complex. (B) Yeast two-hybrid binding assays: PhS11 vs. PhFap7, ScS14a vs. ScFap7, PhFap7 vs. ScS14a, PhS11ΔC15 vs. PhFap7. (C) Fluorescence quenching of fl-PhFap7 upon titration with PhS11 for the determination of the KD of the PhFap7:PhS11 complex. (D) Model of PhS11 bound to ribosomal RNA based on the structure of the T. thermophila SSU X-ray structure (27). Arginine side chains are shown as sticks and labeled. (E) Changes in fluorescence anisotropy upon titration of fluoresceine-labeled h23-RNA (fl-h23, Left) with PhS11. Addition of PhFap7 (arrow) in an amount equimolar to PhS11 leads to a drop in fluorescence anisotropy to a value similar to that for free fl-h23. (F) Competitive binding of PhS11 to unlabeled PhFap7 and 5′ fluoresceine-labeled h23-RNA observed in fluorescence anisotropy measurements. Formation of the PhS11:fl-h23 complex is only detectable upon saturation of PhFap7 by PhS11 (dashed vertical line). (G) Overlay of 15N-TROSY-HSQC spectra of free 15N-Arg-labeled PhS11 (black, signal assignments are indicated) and bound to unlabeled PhFap7 (red). (H) Protection of the C terminus of PhS11 against trypsin digestion by PhFap7 binding analyzed by SDS/PAGE.