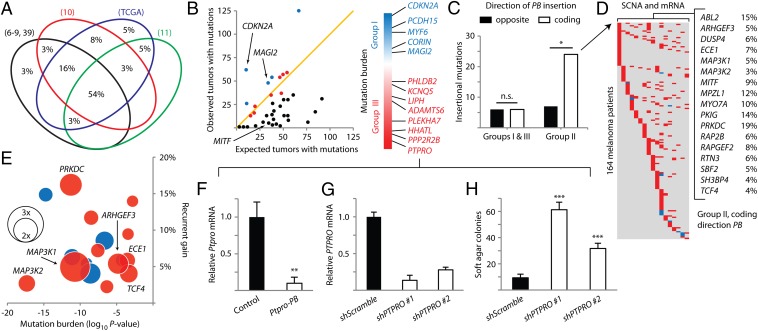
Fig. 4.
PB-implicated genes have a wide spectrum of mutation frequencies in human melanoma. (A) Venn diagram shows the substantial overlap between genes identified in the low-copy PB screen and genes mutated in eight human melanoma mutation datasets (depicted as four ellipses, with the four smallest datasets grouped as one ellipse in black). (B) The detected mutation burden of each gene is plotted against the expected background mutation burden for genes of corresponding sizes. The orange line marks a 1:1 ratio of observed versus background. Genes are colored by group (group I, blue; group II, black; group III, red). Genes in groups I and III are ordered according to the significance of their mutation burdens. (C) The 4× enrichment of coding direction PB insertions is observed in group II genes, predicting overexpression. *P < 0.005, χ2 test. (D) Copy number and expression data for genes enriched in C show significant alterations in 164 out of 222 melanoma patients (amplification/up-regulation in red, deletion/down-regulation in blue). (E) Bubble chart plotting genes from D by significance of mutation burden (x axis, negative values indicate low mutation burden versus background) and percentage of patients with recurrent gains or losses (y axis, positive values indicate percentage of patients with recurrent gain). Expression changes in melanomas relative to normal melanocytes are plotted as bubbles (up-regulation in red, down-regulation in blue). (F) Ptpro expression is diminished in a mouse melanoma harboring insertion in the gene. Relative Ptpro transcript levels in the insertionally mutated melanoma were compared with three melanomas not harboring Ptpro insertion. **P < 0.001, t test. (G) shRNA constructs reduced PTPRO mRNA levels in p’mel-BRAFV600E cells. (H) Depletion of PTPRO in p’mel-BRAFV600E cells promoted soft agar colony formation. Colonies greater than 100 μm in diameter are quantified. ***P < 0.0001, t test. Data in F, G, and H are presented as mean ± SD (n = 3).