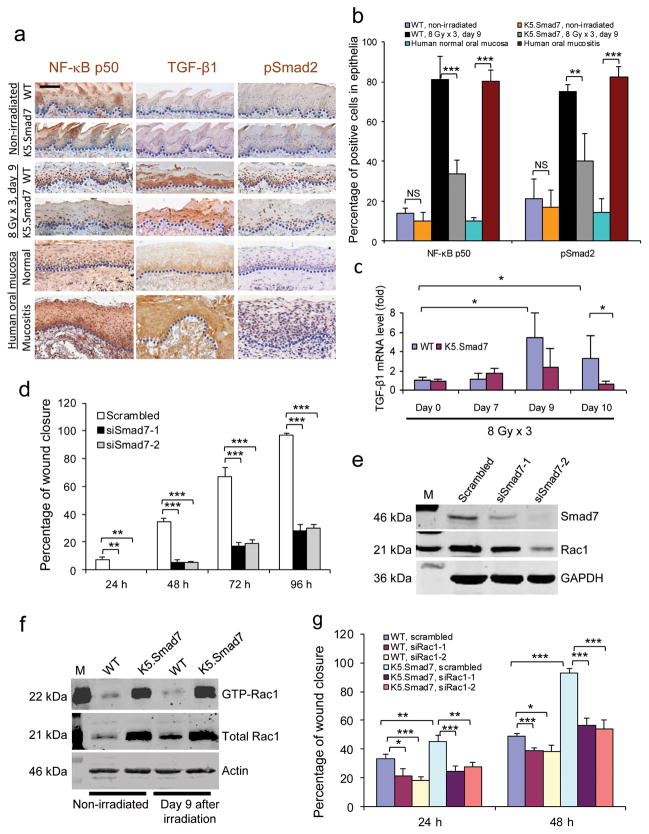
Figure 2. Molecular alterations attenuated by Smad7.
(a) Immunostaining of NF-κB subunit p50, TGF-β1 and pSmad2 in irradiated tongue sections of WT mice adjacent to an ulcer and sections from the damaged area of K5.Smad7 mice, as well as in human samples from nonirradiated oral mucosa and radiation-induced mucositis. Dotted lines delineate epithelial-stromal boundary. Scale bar, 25 μm. (b) Quantification (mean ± s.d.) of nuclear NF-κB subunit p50 and pSmad2 in a. n = 3 or 4 per group. (c) Quantitative RT-PCR (mean ± s.d.) of TGF-β1 (normalized to keratin 5; n = 6 per group for day 0, n = 4 for day 7 and day 9, and n = 7 for day 10). (d) Quantification (mean ± s.d.) of human oral keratinocyte migration (see images in Supplementary Fig. 2). Scrambled, scrambled siRNA; siSmad7-1 and siSmad7-2, two different siRNAs specific to Smad7. n = 3 per group. (e) Western blot analysis of Rac1 72 h after Smad7 knockdown. The knockdown efficiency of siSmad7-1 and siSmad7-2 can be estimated from the blot. M: molecular markers; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (f) Western blot analysis of total and activated (GTP-bound) Rac1 (GTP-Rac1) protein. (g) Effect of Rac1 knockdown on Smad7-mediated keratinocyte migration (see knockdown efficiency in Supplementary Fig. 3a and images in Supplementary Fig. 3d). n = 3 per group. Data are presented as mean ± s.d. siRac-1 and siRac1-2 are two siRNAs specific for Rac1. *P <0.05, **P < 0.01, ***P < 0.001, NS: no significance determined by two-tailed Student’s t test.