Fig. 3.
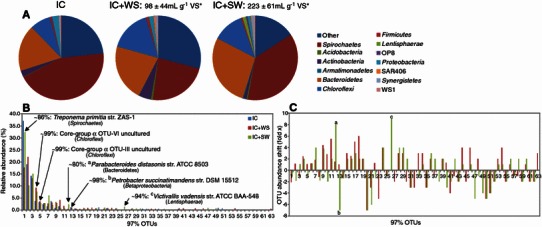
Relative abundance and comparison profiles of bacterial 16S rRNA OTUs identified in anaerobic digesters containing either waste water sludge with no additional organic substrate (inoculum, IC), IC plus wheat straw (IC + WS), or IC plus seaweed (IC + SW). Relative abundance of bacterial lineages at a phylum-level (a) and OTU-level (b) are shown. OTU abundance shifts between WS and SW digesters (c) were measured as either fold-change increases (+) or decreases (−) against IC measurements. Lineage information for selected OTUs (a–c) and OTU affiliation to previously described, highly prevalent core phylotypes (Rivière et al. 2009) is provided. Colour coding in b and c are as follows: IC, red indicates IC + WS and green indicates IC + SW. OTUs numbers in the x-axis corresponds to BAC_nor-terminology referred to in the text. Total methane yields are included in a for IC + WS and IC + SW, which are provided in the original publication on methane production (Vivekanand et al. 2012) and normalized for production in IC. VS* volatile solids
