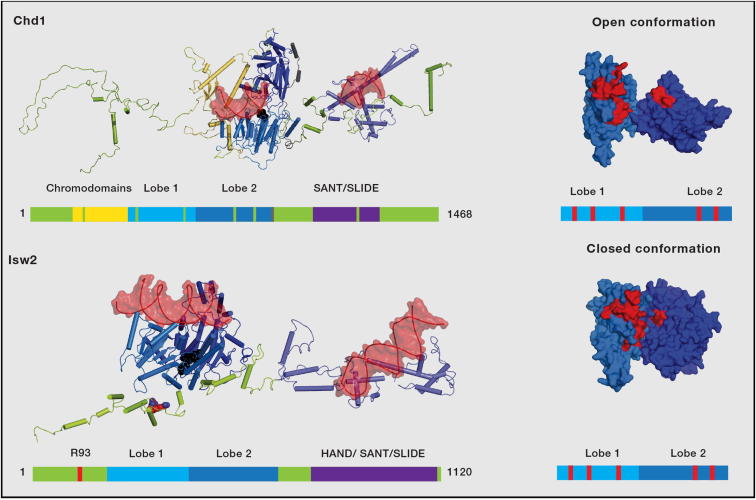
Figure 3.
Structural Models for Chd1 and Isw2
The chromodomains, translocase lobes (Hauk et al., 2010), and SANT/SLIDE DNA-binding domain (Sharma et al., 2011) of Chd1 are colored yellow, blue, and purple, as indicated in the schematic. The structure of linker sequences was crudely modeled based upon secondary structure prediction to indicate their scale rather than conformation. To the right, the helicase lobes are shown as space-fill, with the conserved DNA-binding motifs I, II, and III of lobe I and motifs V and VII of lobe II indicated in red. These conserved motifs are observed to be held in an open conformation that is likely to be inefficient for ATP-dependent DNA translocation. A similar model is shown for Isw2. In this, the HAND-SANT-SLIDE domain is modeled on Isw1 (Yamada et al., 2011), and the ATPase lobes are modeled using the structure of Zebrafish Rad54 in a configuration close to the closed conformation likely to be active for DNA translocation (Thomä et al., 2005). For both Chd1 and Isw2, accessory sequences contribute to the regulation of catalytic activity, and this may well involve changes in the alignment of the ATPase lobes. For example, the chromodomains of Chd1 and R93 of the ISWI protein (in red space-fill) confer negative autoregulation (Clapier and Cairns, 2012; Hauk and Bowman, 2011). This region also undergoes a conformational change upon DNA binding (Mueller-Planitz et al., 2013). In contrast the SANT-SLIDE domains of both proteins confer positive regulation (Hota et al., 2013; McKnight et al., 2011). DNA fragments bound to the SANT/SLIDE domains and modeled into the translocase domain are shown in red space-fill. However, it should be noted that, as the conformation of linker sequences (green) is not known, it is not possible to infer the orientation of the two bound DNA fragments.