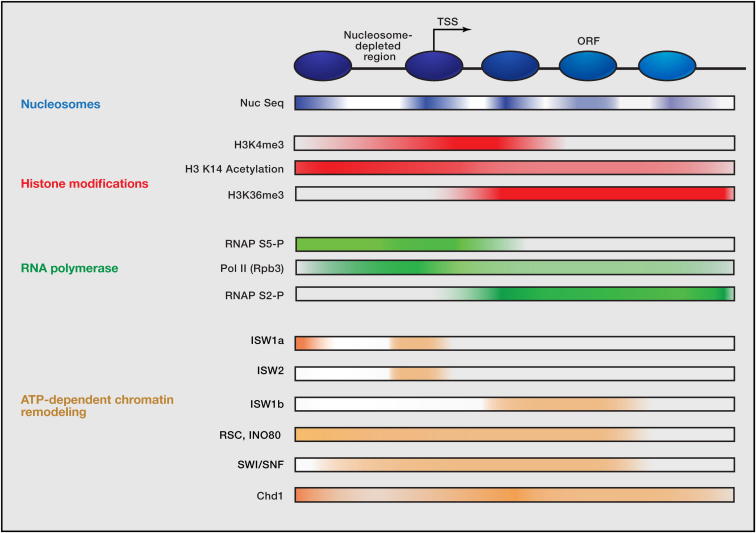
Figure 4.
Organization of Chromatin-Remodeling Enzymes with Respect to Transcribed Genes
A schematic representation organization of chromatin-related factors with reference to the transcriptional start site (TSS). The genome-wide distributions of nucleosomes and posttranslational modifications to histones and RNA polymerase subunits reveal that many of these factors are organized with respect to transcribed genes (reviewed by Rando and Chang, 2009). More recently, it has become apparent that ATP-dependent remodeling enzymes also show distinct distributions with respect to transcribed genes where they influence nucleosome organization (Gkikopoulos et al., 2011; Yen et al., 2012). Furthermore, in some cases, the action of remodeling enzymes can influence the distribution of histone modifications and variants, whereas in other cases, modifications instruct the action of remodelers. This interplay between histone modifications and ATP-dependent chromatin modifications acts to sculpt the chromatin landscape on a genome scale and is likely to involve further integration with transcriptional elongation factors and additional factors acting to regulate chromatin organization, such as histone chaperones.