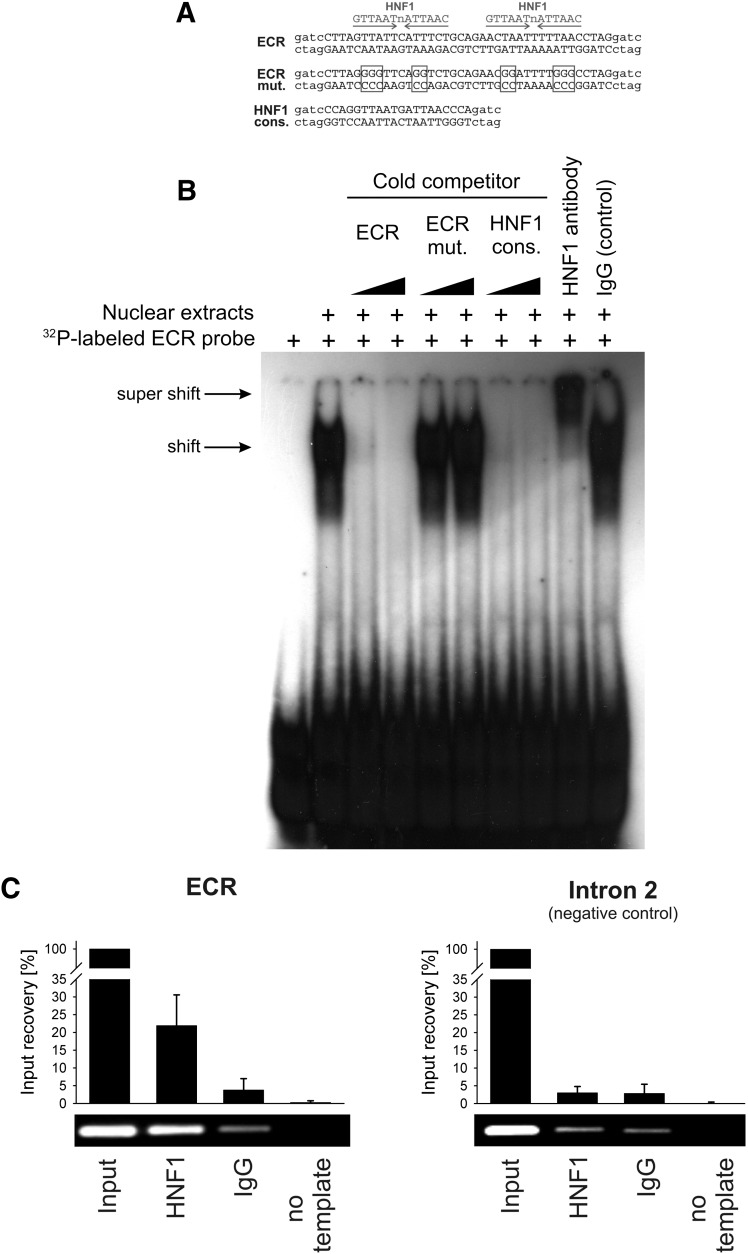
Fig. 2.
Electrophoretic mobility shift and chromatin immunoprecipitation assays demonstrating binding of HNF1 to the evolutionary conserved region in intron 1 of the OCT1 gene. (A) Sequences of the probes used in the electrophoretic mobility shift assay (EMSA). The specific sequences are given in upper case, and the unspecific sequences used in the radioactive labeling of the EMSA probes are given in lower case letters. Positions mutated to disrupt the HNF1 binding sites are indicated by boxes. (B) The 32P-labeled probe containing the 40-bp highly conserved sequence from the intron 1 ECR was incubated with nuclear extracts from HepG2 cells in the absence or presence of unlabeled probes (cold competition) or antibodies (supershift). The unlabeled probes were given in 3- and 30-fold molar excess of the 32P-labeled probe. (C) Chromatin immunoprecipitation assay (ChIP) of isolated human hepatocytes. A representative agarose gel and a real-time PCR-based signal quantification are shown. The intron 1 ECR and a negative control region in intron 2 of the OCT1 gene were analyzed. The quantification results are based on three independent ChIP experiments quantified in duplicate and are shown as means and standard deviations. Identical HNF1-specific and control IgG antibodies were used in the electrophoretic mobility shift and chromatin immunoprecipitation assays. Detailed information about the antibodies used is available in the text.