Figure 3. The Long Displaced ssDNA Tail and MutS tetramerization are Involved in MutS-MutL-Imposed Antirecombination.
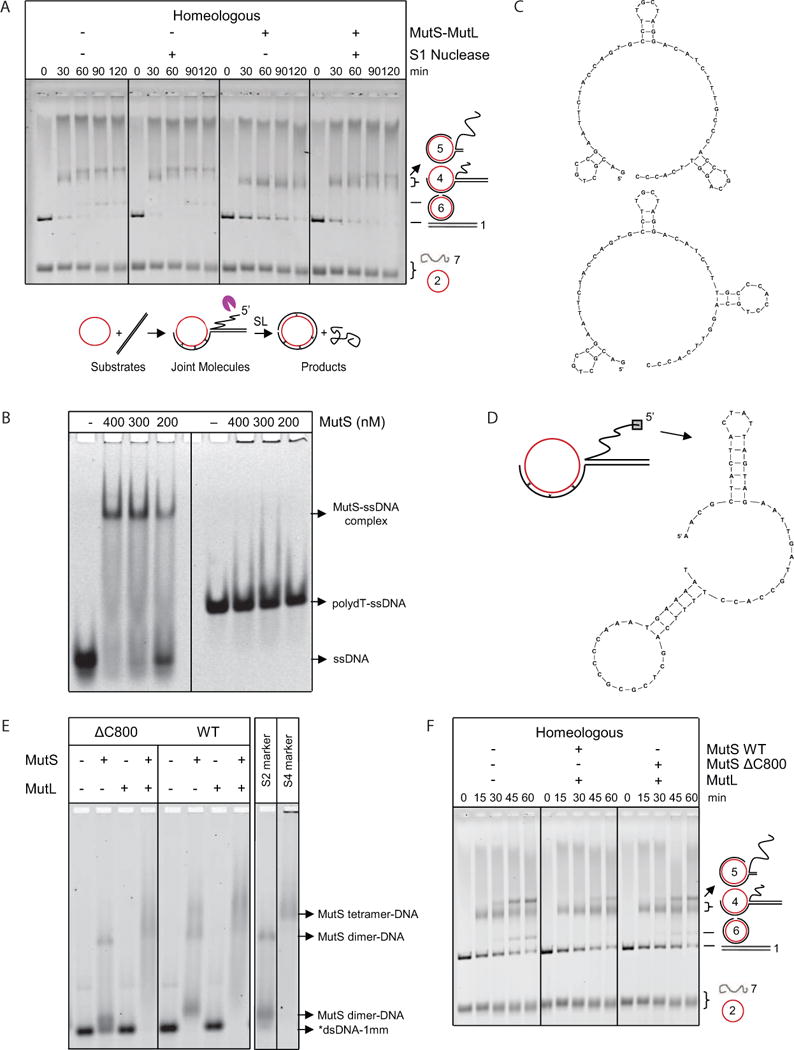
(A) S1 nuclease-mediated degradation of the 5′ displaced ssDNA reduces inhibition of homeologous strand exchange by MutS-MutL. Homeologous strand exchange reactions from left to right: control; with S1 nuclease (0.04 U/μl); with MutS-MutL (100, 100 nM); with S1 nuclease (0.04 U/μl) and MutS-MutL (100, 100 nM). Substrates, intermediates and products are shown below the gel. The purple pacman symbol represents the S1 nuclease. See also Figure S3.
(B) Electrophoretic mobility shift assay of increasing concentrations of MutS binding to ssDNA containing secondary structures and poly-dT.
(C) Predicted secondary structures formed in the ssDNA with arbitrary sequence used in panel B.
(D) Predicted secondary structures formed in the first 60 nucleotides of the 5′ displaced strand of M13.
(E) Binding of wild type MutS, ΔC800 MutS and MutL (400 nM each) alone and together to labeled 90-bp dsDNA with 1 mismatch (*dsDNA-1mm). Markers for DNA-bound MutS dimer (S2) and tetramer (S4) (Groothuizen et al., 2013) are indicated in the panel on the right.
(F) In the presence of MutL, MutSΔC800 only partially blocked homeologous strand exchange as indicated by the formation of higher amounts of DNA intermediate 5 and product compared to the reaction with wild-type MutS-MutL. From left to right, homeologous strand exchange reactions without MutS-MutL, with MutS-MutL (75, 75 nM) and with MutSΔC800-MutL (75, 75 nM).
