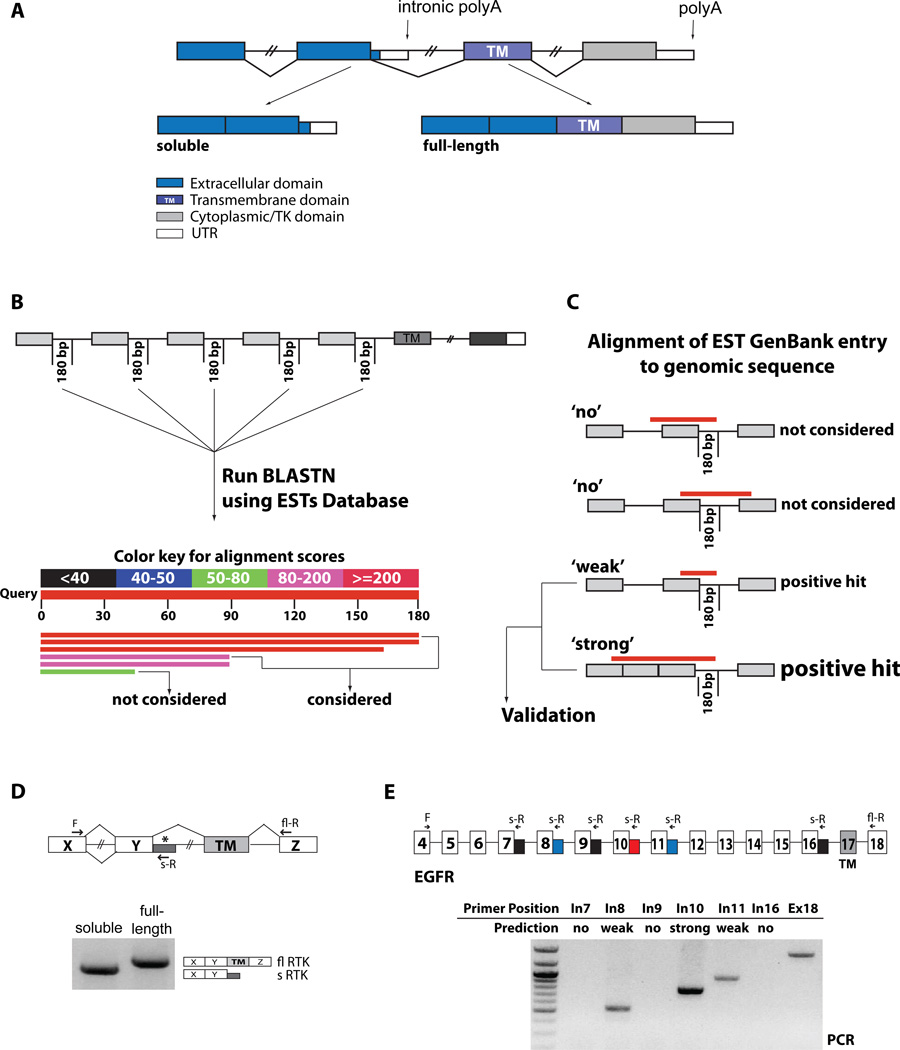
Figure 1. Identification and validation of soluble RTK isoforms.
(A) Alternative polyadenylation in RTKs. Alternative PAS usage upstream of the TM domain leads to soluble isoforms that compete as decoy receptors with the membrane-anchored, full-length isoforms. (B) The first 180 bps of selected introns were used to query EST databases with BLASTN. Positive hits (score>80) were further analyzed. (C) Retrieved GenBank EST hit entries were aligned to their genomic sequences, and hits that crossed the upstream exon-intron boundary were pursued. Hits extending into the upstream intron, reflecting unprocessed RNA or genomic contaminations, were discarded. Hits that reflected full intron retention were also discarded since these variants would still generate soluble variants, but by a different, NMD-sensitive mechanism. Hits that ended within the first upstream exon were considered ‘weak’ since neither upstream processing nor unprocessed RNA can be excluded. Hits that aligned to the exon-intron boundary and spanned one or more upstream exon-exon junctions, indicating evidence of processed RNAs were considered ‘strong’ positive hits. All weak and strong positive hits were further evaluated for validation by oligo-dT RT-PCR. (D) PCR validation was performed with forward primers located on exons upstream of the one preceding the intronic hit, to ensure that the amplified product derives from processed mRNAs. Reverse primers were located within the intronic hit (soluble RTK) or within an exon downstream of the TM domain (full-length) as a measure of expression of the gene. The products of the PCR reaction were analyzed by gel electrophoresis and sequenced. (E) RT-PCR validation of predicted soluble EGFR variants. Reverse primers were designed to amplify the predicted new 3’UTRs (blue and red) from spliced mRNAs. Reverse primers within introns without predicted EST hits were used as negative controls (black). A common forward primer sequence is located in exon 4, the full-length reverse primer is in exon 18. See also Supp. Table s1.