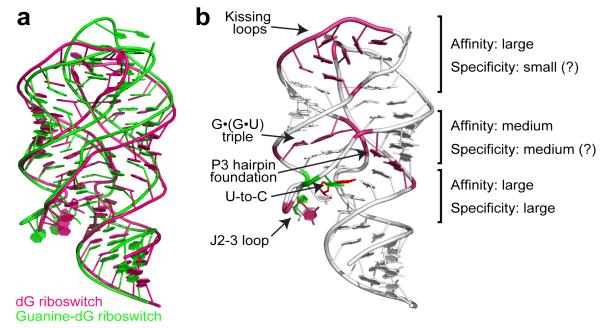
Figure 6.
Summary of dG recognition. a, Superposition of the natural dG riboswitch structure (this study, pink) and the guanine riboswitch based guanine-dG riboswitch structure17 (green) shows similar global RNA folds. b, Involvement of riboswitch structural features in dG binding to the natural dG riboswitch. Pink and green nucleotides designate areas where the most critical differences from adenine/guanine riboswitches occur. Green nucleotides highlight similarities with the guanine-dG riboswitch structure17. Each element’s contribution to the specificity and affinity of dG binding is roughly estimated based on current and published data13,17.