Abstract
Whole genome sequencing has allowed rapid progress in the application of forward genetics in model species. In this study, we demonstrated an application of next-generation sequencing for forward genetics in a complex crop genome. We sequenced an ethyl methanesulfonate-induced mutant of Sorghum bicolor defective in hydrogen cyanide release and identified the causal mutation. A workflow identified the causal polymorphism relative to the reference BTx623 genome by integrating data from single nucleotide polymorphism identification, prior information about candidate gene(s) implicated in cyanogenesis, mutation spectra, and polymorphisms likely to affect phenotypic changes. A point mutation resulting in a premature stop codon in the coding sequence of dhurrinase2, which encodes a protein involved in the dhurrin catabolic pathway, was responsible for the acyanogenic phenotype. Cyanogenic glucosides are not cyanogenic compounds but their cyanohydrins derivatives do release cyanide. The mutant accumulated the glucoside, dhurrin, but failed to efficiently release cyanide upon tissue disruption. Thus, we tested the effects of cyanide release on insect herbivory in a genetic background in which accumulation of cyanogenic glucoside is unchanged. Insect preference choice experiments and herbivory measurements demonstrate a deterrent effect of cyanide release capacity, even in the presence of wild-type levels of cyanogenic glucoside accumulation. Our gene cloning method substantiates the value of (1) a sequenced genome, (2) a strongly penetrant and easily measurable phenotype, and (3) a workflow to pinpoint a causal mutation in crop genomes and accelerate in the discovery of gene function in the postgenomic era.
Keywords: Bioinformatics, Illumina, mutation discovery, forward genetics
THE advent of high throughput, short read sequencing (also known as next-generation sequencing, NGS) and the availability of whole genome sequences can accelerate the molecular study of genetic phenomena. In genetic model organisms with small genome sizes, such as Arabidopsis thaliana, genes disrupted by ethyl methanesulfonate (EMS) mutations have been cloned by shotgun sequencing of DNA derived from pooled mutants (Schneeberger et al. 2009; Austin et al. 2011; Mokry et al. 2011; Uchida et al. 2011; Lindner et al. 2012; Zhu et al. 2012a,b) analogous to the bulked segregant analysis approach (Michelmore et al. 1991). Community efforts to generate crop genome sequences were expected to enable researchers to quickly clone genes to answer biologically relevant and agriculturally pertinent questions. NGS-enabled cloning has been successful in maize for cloning transposon insertion sites (Williams-Carrier et al. 2010) and in rice for agronomically important traits (Abe et al. 2012).
Sorghum is a cereal crop of critical importance as a food and forage source in hot, arid environments and is a promising bioenergy crop in the United States. The sorghum genome of accession BTx623 is sequenced and well annotated (Paterson et al. 2009). It is a diploid species with a genome size of 700 Mb, roughly five times the size of A. thaliana. Sorghum is genetically variable with many diverse genetic stocks including natural (Casa et al. 2008) and induced variation (Blomstedt et al. 2012). This sequenced genome makes sorghum an excellent system to demonstrate the practicality of NGS for forward genetics and mutant gene identification. The utility of sequencing mutants to identify the causative polymorphism was tested using a metabolite phenotype connected to insect herbivory.
Cyanogenic glycosides (CGs) belong to a class of plant allelochemicals which negatively impact animal and human nutrition. More than 2650 plant species are cyanogenic and release hydrogen cyanide (HCN) upon tissue disruption (Conn 1981; Seigler and Brinker 1993), making the plant material toxic to humans and most animals (Oluwole et al. 2000). Given the toxicity of HCN, CGs are known to play a role in plant defense against herbivores (Scriber 1978; Glander et al. 1989; Mowat and Clawson 1996; Ferreira et al. 1997; Tattersall et al. 2001). In sorghum, genes encoding the enzymes for synthesis and catabolism of the predominant CG, dhurrin, have been determined (reviewed in Carlson 1958; Ganjewala and Kumar 2010). Dhurrinase1 (dhr1) and dhurrinase2 (dhr2) are enzymes responsible for HCN release in sorghum and exhibit high specificity for their physiological substrate, dhurrin (Hösel et al. 1987). Dhr1 accumulates in the mesocotyl and in root tips, while dhr2 accumulates in leaves (Thayer and Conn 1981; Hösel et al. 1987; Cicek and Esen 1998). It is speculated that HCN release following tissue damage by chewing deters insect herbivory of sorghum (Jones 1962; Hughes 1991; Schappert and Shore, 1999, 2000; Gleadow and Woodrow 2002). Thus far, a single acyanogenic sorghum mutant, deficient in dhurrin biosynthesis due to loss of function of CYP79A1, has been described (Blomstedt et al. 2012) but many more genes are known to be involved in dhurrin biosynthesis and catabolism. Genetic lesions responsible for acyanogenic sorghum mutant phenotypes should be identifiable using (1) the sequenced genome of sorghum, (2) current NGS technology, (3) an easily quantifiable phenotype for HCN production, and (4) an expertly curated candidate gene list.
In this study, we used NGS to identify a mutation in the dhurrin catabolic pathway that is the cause of an acyanogenic sorghum mutant phenotype. A high-throughput colorimetric technique was used to screen a sorghum EMS-mutagenized population for acyanogenic individuals. Whole genome, short read sequencing of one acyanogenic sorghum mutant individual identified a single EMS mutation on chromosome 8 predicted to truncate the dhurrinase2 protein, a catabolic enzyme that is required for dhurrin deglucosylation and subsequent HCN release. The phenotype was demonstrated to cosegregate with the polymorphism and to alter insect choice and herbivory. Despite the discovery of dhurrin and with it the requirement for dhurrinase enzyme activity in 1902 (Dunstan and Henry 1902), this is the first report of a mutant defective in dhurrinase activity.
Materials and Methods
Cultivation of plants
Prior to sowing, sorghum [Sorghum bicolor (L.) Moench] seeds were treated with a suspension of Maxim 4FS (0.08 ounces, oz.), Apron XL (0.32 oz.), colorant (0.25 oz.), and water (12.7 oz.). Treated seeds were grown in the Purdue University Agronomy greenhouse facilities and either sown in sand benches or in 2-gallon plastic pots and flats filled with a 1:1 mix of sand and local soil (sandy loam, 2.0% organic matter) as substrate. Growth conditions were as follows: 14 hr light period with 21–33°. Plants were fertilized with Osmocote (14-14-14; Scotts, Marysville, OH).
Identification of an acyanogenic sorghum line
Three sets of ∼1.5 kg BTx623 seeds were treated with 25, 35, or 45 mM EMS for ∼20 hr and then rinsed thoroughly with water. The mutagenized seeds were planted at the Purdue Agronomy Center for Research and Education (ACRE, West Lafayette, IN) in the summer of 2009. The 45-mM rate of EMS produced M1 plants with the most severe chemical injury (data not shown). Selfed families from 1000 fertile plants treated with 45 mM EMS were used for a mutant screen.
We screened the EMS population for variation in HCN release using the Feigl–Anger paper assay as previously described (Hay-Roe et al. 2011). This test measures HCN release capacity (HCNc) (Alonso-Amelot and Oliveros-Bastidas 2005; Ballhorn et al. 2005). Approximately 15 to 20 seeds were grown from 1000 of the EMS M2 families and 300 genotypes of the sorghum association panel. From each genotype, the youngest leaf was sampled from 10 individuals at the 2- to 3-leaf stage and placed in 96-well plates. Plates were then incubated at –80° for 3 hr. The plates were thawed, wiped clean of moisture with paper towels, and covered with Feigl–Anger paper (Feigl and Anger 1966; Kakes 1991; Takos et al. 2010; Blomstedt et al. 2012). A 96-well plate lid was placed over the paper to provide a tight fit. Plates were incubated at 35° for ∼20 min to allow tissue lysis. Blue spots on Feigl–Anger paper indicate HCN release and an absence of a blue spot indicates a lack of HCN release and an acyanogenic phenotype (Supporting Information, Figure S1).
HPLC determined dhurrin content in the youngest fully expanded leaf from seedlings and adult sorghum. Fresh weight was recorded. Samples were immersed in 500 µl 50% methanol at 75° for 15 min to inactivate catabolic enzymes and extract metabolites. Samples were analyzed on a Shim-pack XR-ODS column (3.0 × 75 mm, 2.2 μm; Shimadzu, Kyoto, Japan) using a gradient from 5% acetonitrile in 0.1% formic acid to 15% acetonitrile in 0.1% formic acid at a flow rate of 1 ml min−1. Dhurrin was quantified by using p-hydroxybenzaldehyde as a standard (De Nicola et al. 2011).
Next generation whole genome sequencing
DNA isolation for NGS sequencing was conducted by grinding 100–200 mg of young leaf tissue in a 1.5-ml microfuge tube in liquid nitrogen using a metal rotary pestle and electric grinder. To each tube, 700 μl of extraction buffer [7 M urea, 50 mM Tris-HCl pH 7.5, 0.3 M NaCl, and 20 mM EDTA pH 8.0, 2% (w/v) Sarkosyl] was added. Then, 700 µl of phenol:chloroform:isoamyl alcohol (25:24:1) saturated with 10 mM Tris pH 8.0 (Sigma-Aldrich, St. Louis) was added. After thorough mixing, the sample was centrifuged at 17,950 × g (Centrifuge 5424 Eppendorf; Hauppauge, NY) for 10 min. The aqueous phase was then transferred to a clean 1.5-ml microfuge tube. DNA was precipitated by adding 1/10 volume sodium acetate (pH 5) and then an equal volume of cold isopropanol. DNA was pelleted by centrifugation (17,950 × g) for 10 min, washed with 70% ethanol, and resuspended in 200 µl of distilled water.
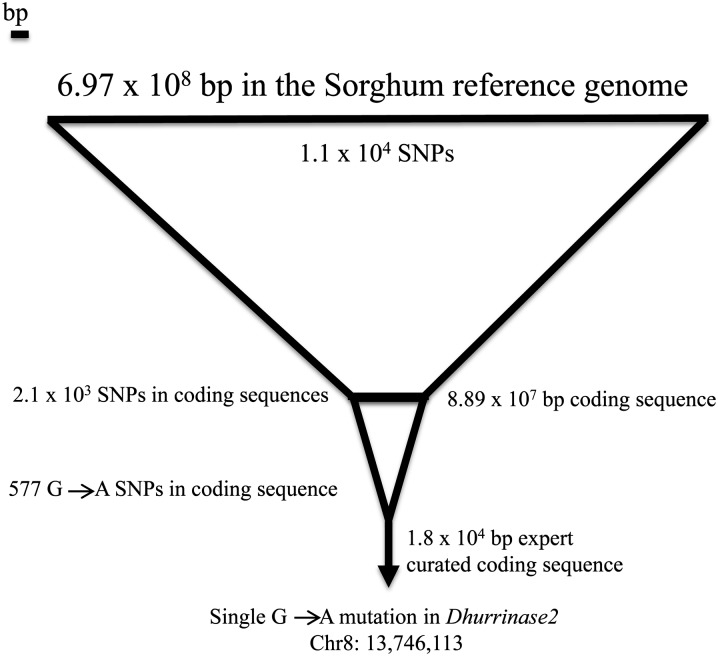
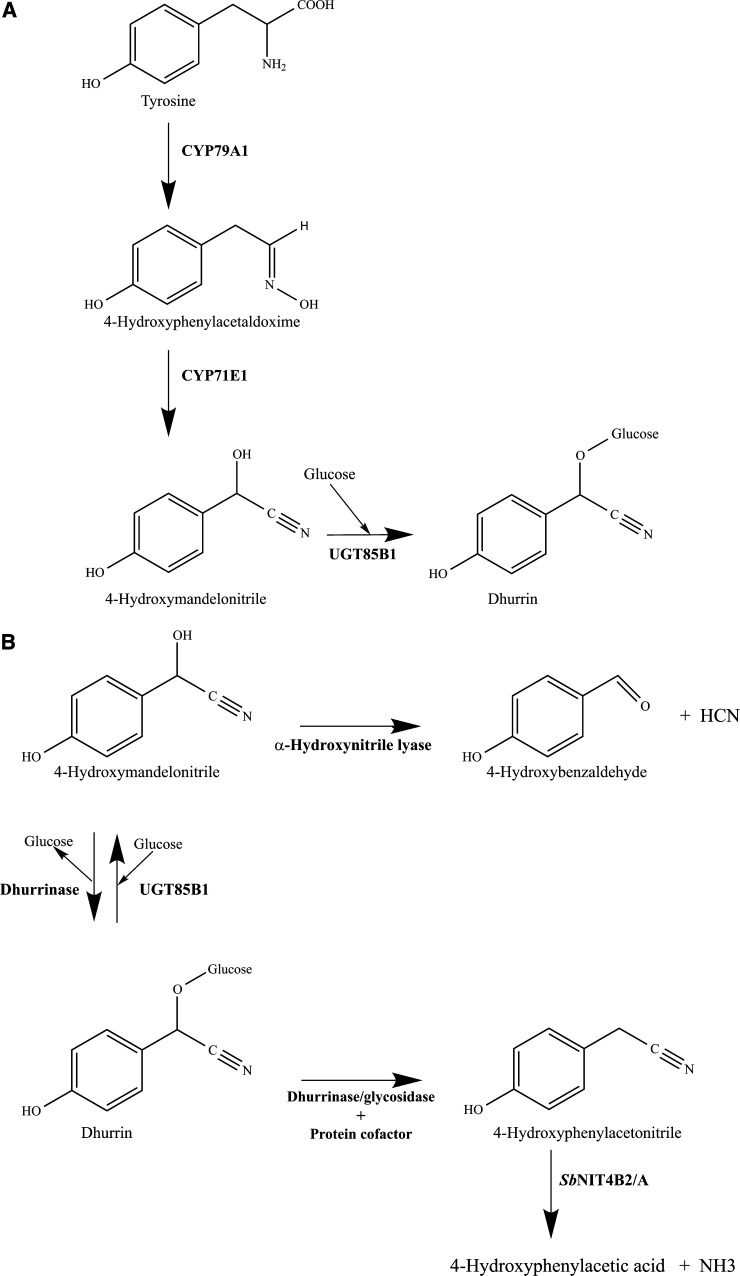
To identify EMS mutations in the acyanogenic mutant, the genome of a single homozygous mutant plant was sequenced using an Illumina HiScanSQ instrument (Illumina, San Diego) at the Purdue University Genomics Core Facility (West Lafayette, IN). Approximately 20 million 100-bp paired-end sequencing reads resulted in 17-fold genomic coverage when aligned to the BTx623 reference genome sequence (Paterson et al. 2009). Single nucleotide polymorphisms (SNPs) were called using the Illumina CASAVA pipeline (software version 1.7.0, 201l). The sorghum genome includes nearly 7 Mb of assembled DNA unassigned to any of the 10 chromosomes. We focused on the SNPs within the assembled 10 chromosomes. To identify the EMS mutant within this collection of real and false-positive SNP calls, a workflow was created to identify the gene(s) responsible for any EMS mutant (Figure 1). SNPs were filtered by the following criteria: (1) Coverage must be <100 reads to remove many repetitive elements; (2) SNPs must be identified as homozygous; (3) SNP-containing reads must be derived from both strands; (4) SNPs must fall within the coding regions of the sorghum genome; and (5) EMS mutations cause G-to-A nucleotide changes (Henikoff et al. 2004), so only G:C to A:T transitions were retained. All filtering was done using Galaxy (Blankenberg et al. 2001; Goecks et al. 2010; https://main.g2.bx.psu.edu/). The annotation of the genome contains three genes from the dhurrin biosynthetic pathway (CYP79A1, CYP71E1, and UGT8581; Figure 2A) and three genes from the catabolic pathway (dhurrinase1, dhurrinase2, and alpha-hydroxynitrile lyase; Figure 2B; Table S1).
Figure 1.
NGS SNP narrowing to causative mutation. The lines are representative of base pairs (bp) of the genome. The coding sequence represents ∼13% of the whole genome. Steps for SNP filtering are as follows: (1) SNPs within the coding sequences, (2) G-to-A as well as complementary C-to-T SNPs in coding sequence, and (3) SNPs within the candidate gene list.
Figure 2.
Metabolism of cyanogenic glucosides in sorghum [Sorghum bicolor (L.) Moench]. (A) The biosynthesis of cyanogenic glycosides in sorghum is catalyzed by three enzymes CYP791A, CYP71E1, and UGT85B1. (B) The cyanogenic glucosides in sorghum are catabolized by two enzymes, dhurrinase and α-hydroxynitrile lyase. Dhurrin turn-over is supposed to involve a dhurrinase and a protein cofactor.
Mutation confirmation by Sanger sequencing and cosegregation analysis
PCR amplification conditions for the dhr2 gene were as follows: (step 1) initial incubation at 95° for 3 min; (step 2) 34 cycles of 95° for 30 sec, 55° for 30 sec, and 72° for 1 min; and (step 3) 72° for 5 min. Primers used were: Dhr2F1 (5′-AACTTGTTGATAGACAACGGCATA-3′) and Dhr2R1 (5′-AAGGTATGGCTCTCTGATTGAGTC-3′) (Nielsen et al. 2006). A Zymoclean gel DNA recovery kit (Zymo Research, Irvine, CA) was used to clean and recover the PCR products. Sanger sequencing of the PCR products was performed at the Purdue Genomics Core Facility.
A KASPar genotyping assay was used to assess cosegregation between the dhr2-1 EMS allele and the HCNc phenotype in a segregating F2 population created by crossing the BTx623 parent with the mutant. The following sequence was used to generate a KASPar (Kbioscience, Hoddesdon, UK) assay for SNP detection (CATTATCCAATGCGCAAATTTAAGTTTTGACGAATCATTCTTAATTTACTCATTGCAACTCATCAGGCATAGAGCCATATGTAACAATTTTCCACTG(G/A)GACACGCCTCAAGCACTGGTAGATGACTACGGCGGCTTCTTAGATAAGAGAATTATGTAAGTACTACTCCCTCCATCCCAAATTGTAAGTCATTCCAAGAATCTTGGAGAGTCAAAGCA). DNA was extracted from a segregating F2 population. Approximately 40 ng of DNA, 2 µl of the 2X KASPar reaction mix and 0.055 µl of KASPar assay (Kbioscience) were used per reaction. PCR conditions were as follows: (step 1) initial incubation at 94° for 15 min; (step 2) 10 cycles of 94° for 20 sec, followed by 60 sec at 65° in the first cycle dropping 0.8° per cycle with the final cycle at 57°; (step 3) 94° for 20 sec, 57° for 60 sec repeated for 26 cycles on a Roche Light cycler 480 (Roche Applied Science, Indianapolis). Fluorescence from the PCR product was read using the Roche Light cycler 480 (Roche Applied Science) and the data analyzed using the LightCycler 480 SW 1.5 software program (Roche Applied Science).
Insect herbivory measurements
Insect feeding choice of fall army worm (Spodoptera frugiperda) was observed on the acyanogenic sorghum. S. frugiperda eggs and third instar larvae were obtained from Benzon Research (Carlisle, PA). Eggs were hatched in a 1-gallon plastic bag with high humidity at 29° in an incubation chamber. The hatched larvae were then transferred into feeding media containers (Benzon Research) and allowed to grow to third through sixth instar at 28°.
S. frugiperda settling preference choice experiments compared wild type (BTx623) to the acyanogenic mutant. The youngest, fully expanded leaves from 3-wk old BTx623 sorghum wild-type and mutant plants were harvested. Three leaves from a genotype were bundled into a stack and five stacks including at least two of each genotype were placed around the perimeter of plastic containers in a randomized design. The third through sixth instar stage S. frugiperda larvae were starved for 3 hr and released into the center of the container. Insects were allowed to feed for 12 hr and the settling preference of the insects was measured by counting the number of insects on each of the leaf stacks.
Whole plant insect settling preference as well as the S. frugiperda feeding habits were characterized over a 24-hr period. The BTx623 parent and the acyanogenic mutant plants were grown in 2-gallon pots on a 1:1 mix of sand and soil. Ten 3-wk-old plants of each genotype were transplanted into plastic flats and were randomly placed. The third through sixth instar stage S. frugiperda larvae were starved for 3 hr and released into the center of the flats at the rate of 60 insects per flat. The entire flat, along with the plants and insects, were covered with a transparent plastic dome to prevent insect escape. Twenty-four hours after infestation, the settling preference of the insects was measured by counting the number of insects on each plant. The similarity between the preinfested fresh weights of the plants of both parent and mutant genotypes was established by performing a t-test on the fresh weights of randomly selected plants from both genotypes of each plastic flats. Postinfested fresh weight measurement of plants of both the genotypes was done 24 hr after insect infestation.
Results
Identification of an acyanogenic mutant of S. bicolor
To find sorghum compromised in HCN release, an EMS-treated population of BTx623 was analyzed for HCN release (HCNc) using the Feigl–Anger paper method (Feigl and Anger 1966; Kakes 1991; Takos et al. 2010; Hay-Roe et al. 2011; Blomstedt et al. 2012). Approximately 1000 independent M2 families were screened. From this screen, a single family was identified with no Feigl–Anger detectable HCNc following incubation at 35° (Figure S1). We refer to this slow HCN emitting phenotype as “acyanogenic” for simplicity. The slow HCN emitting plants were indistinguishable at a gross morphological level from the wild type (Figure S2). Flowering time and seed set produced by the mutant were also similar to the wild-type parent (data not shown). Given the apparent lack of a phenotype for HCNc suppression, we tested 300 sorghum inbreds to see if a natural variant could be identified in which HCNc was similarly decreased. A selection of 300 genotypes from the sorghum association panel (Casa et al. 2008) from diverse geographic and genotypic backgrounds was analyzed by the Feigel–Anger paper assay. HCNc varied among the different accessions but no null phenotypes were observed (Figure S1).
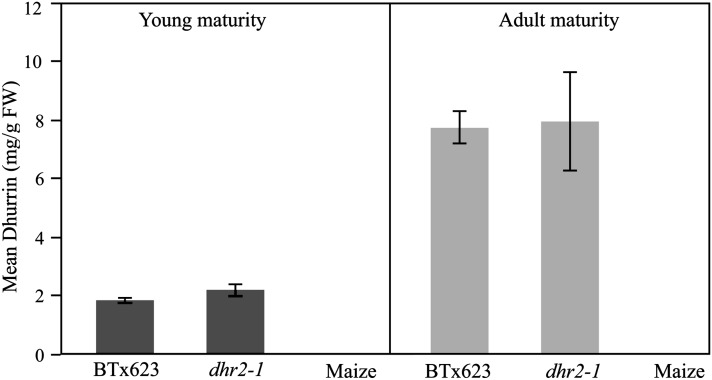
Disruption in the dhurrin biosynthetic or catabolic pathways would block HCNc. Dhurrin content of the acyanogenic mutant and BTx623 plants at different growth stages was quantified by HPLC. Maize (Zea mays) does not produce dhurrin and was used as a negative control. As shown in Figure 3, the HCNc mutant plants had dhurrin contents similar to wild-type BTx623, suggesting that dhurrin biosynthesis was unaffected. A reduction in dhurrin between the seedling and adult plants was observed in both BTx623 wild type and the mutant (Figure 3), consistent with previous observations (Dunstan and Henry 1902; Akazawa et al. 1960; Conn 1994). Given that dhurrin accumulated in the mutant, the acyanogenic phenotype is likely due to a disruption of dhurrin catabolism.
Figure 3.
HPLC analysis was performed to estimate dhurrin content of young seedlings and adult sorghum BTx623 and dhr2-1 mutant plants (mg of dhurrin per g of fresh weight). Similarly developed maize leaves were used as negative controls. Along the x-axis is the dhurrin content and the y-axis represents the mean value of either adult or seedling leaf tissue from sorghum BTx623, dhr2-1 mutant, or maize.
Identification of the dhurrinase2-1 mutation by NGS of acyanogenic sorghum
A NGS sequencing approach was used to identify the causal mutation of the acyanogenic sorghum phenotype. We sequenced a homozygous acyanogenic mutant plant. Over 20 million 100-bp reads, ∼2 million reads per chromosome, were obtained from the mutant and aligned to the reference genome (BTx623; Paterson et al. 2009) using the CASAVA pipeline (2011, v1.7.0). There are 700 (6.97 × 108) Mb in the Sorghum genome. Approximately 11,625 SNPs were called between the reference genome and the acyanogenic mutant. Two different comparisons were carried out to narrow the causative SNP location. Of the 11,625 SNPs, 2131 SNPs were within coding sequence. As the vast majority of EMS mutations are G:A transitions (Henikoff et al. 2004), we further limited our search to G:A, and the complementary C:T, SNPs, leaving 577 SNPs (Figure 1). Simultaneously, a set of six candidate genes was identified representing the dhurrin biosynthetic and catabolic pathways (Figure 1). Using the six candidate genes, the 700 Mb sorghum genome of nearly 30,000 coding sequences or ∼13% (8.89 × 107 bp) of the genome was narrowed to <0.02% (1.8 × 104 bp) of the genome. Three enzymes are involved in dhurrin catabolism (Hösel et al. 1987; Wajant and Mundry 1993) and currently, four genes have been annotated as dhurrinase-like (Sb08g007610, Sb08g007650, Sb08g007586, and Sb08g007570) and one gene (Sb04g036350) as hydroxynitrile lyase-like in the sorghum genome (Paterson et al. 2009; http://www.phytozome.net). A single G-to-A SNP transition was identified in the Durrinase2 (dhr2) (Sb08g007610) gene. This transition results in a nonsense mutation (Tryptophan 194 to STOP) within the first third of the protein (Figure 4) and is predicted to cause a nonfunctional enzyme. We have named this mutant allele, dhr2-1, to match the underlying molecular identity and nomenclature of the disrupted gene. To confirm the EMS-induced mutation, we used targeted Sanger sequencing (Purdue Genomics Core facility) of PCR products from DNA isolated from dhr2-1 mutants and BTx623 (data not shown). The G-to-A nonsense SNP was observed in the mutant but not the wild-type allele, which confirmed the previously identified SNP.
Figure 4.
Protein sequence alignment of durrinase2 (dhr2) (Sb08g007610) from cDNA translation and the projected dhr2 protein truncation. In the dhr2-1 mutant, the tryptophan 194 is converted to a stop codon due to the single G-to-A transition. Shading represents the projected glycosyl hydrolase domain (Marchler-Bauer et al. 2011; http://www.ncbi.nlm.nih.gov). Amino acids in boldface type represent shared motifs between cyanogenic glucosides (Cicek et al. 2000).
Cosegregation of the dhr2-1 allele and HCNc
To confirm linkage of the putative causal mutation in dhr2 to the acyanogenic phenotype, we tested cosegregation of the nonsense SNP in Sb08g007610 and the HCNc phenotype. F1 plants from a cross between BTx623 and dhr2-1 mutants all exhibited the typical rapid blue spot development phenotype associated with the BTx623 parent on Feigl–Anger paper (Feigl and Anger 1966; Kakes 1991; Takos et al. 2010; Hay-Roe et al. 2011; Blomstedt et al. 2012), indicating that dhr2-1 is recessive to the wild-type dhr2 allele. A KASPar (Kbioscience) assay was developed using the G-to-A nonsense SNP in Sb08g007610 and applied to the F2 segregating population. The segregation ratio for this codominant molecular marker was consistent with the Feigl–Anger assay (data not shown). Of 25 HCNc negative plants screened, all were identified as dhr2-1 homozygotes on the KASPar assay (Table S2). Thus, the acyanogenic phenotype in dhr2-1 is a recessive mutation at a single genetic locus and the G-to-A SNP in dhr2 is the likely causal mutation.
Insect herbivory measurements of acyanogenic sorghum
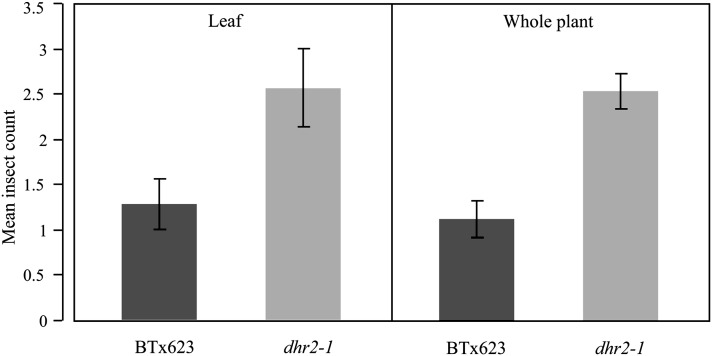
Settling preference choice tests of S. frugiperda, the fall army worm, were conducted to evaluate the role of HCN release in deterrence of insect herbivory. One settling preference test was conducted with sorghum leaf stacks in which S. frugiperda larvae were allowed to choose between sorghum leaf cuttings from the BTx623 and dhr2-1 genotypes. Eighteen independent measurements were made for each genotype. Settling preference of the insects was quantified by counting the number of insects present on each genotype 12 hr after infestation. A two-tailed t-test (JMP; v8.0.1) indicated that a greater number of insects settled on the dhr2-1 acyanogenic mutant leaf cuttings than the BTx623 wild-type leaf cuttings (P < 0.0175; Figure 5).
Figure 5.
Insect (S. frugiperda) settling preference for BTx623 wild-type sorghum and dhr2 leaf tissue. Five replicates of randomly placed leaf stacks of young leaflet cuttings or whole plants of BTx623 and dhr2-1 genotypes. The settling preference of the insects was measured by counting the number of insects on each of the leaf stacks 12 hr after infestation or 24 hr after infestation for the whole plant experiment.
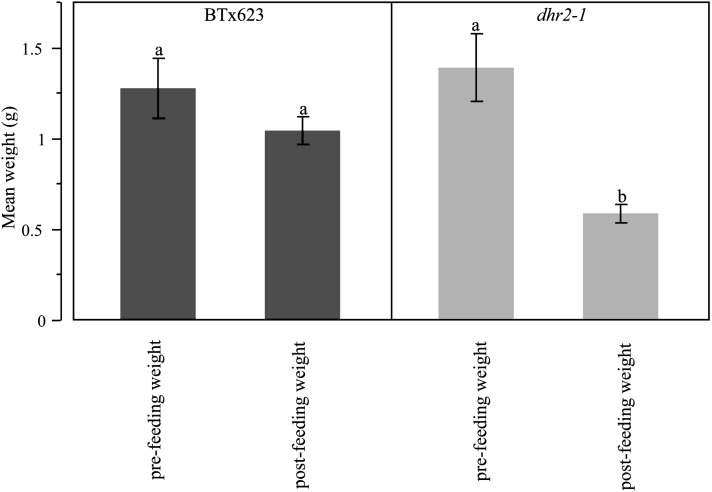
A second settling preference test was conducted with whole plants in greenhouse trays. In this experiment, infestation counts as well as pre- and postinfestation weights were measured in 17 independent tests for each genotype. As with the leaf cutting settling experiment, a t-test indicated a significantly higher number of insects settled on the dhr2-1 mutant plants compared to the BTx623 parent plants (P < 0.001; Figure 5). S. frugiperda both chose acyanogenic mutant individuals and consumed them to a greater degree than the wild-type plants (Figure 6). A t-test failed to detect a difference between fresh weights of the BTx623 and dhr2-1 plants prior to infestation. Similarly there was no difference between controls and postinfestation weights within the wild-type plants (P > 0.3276; Figure 6). Increased herbivory of the dhr2-1 acyanogenic mutants resulted in differences in postinfestation fresh weight in comparisons. Both the comparison between BTx623 and dhr2-1 postinfestation weights and the comparison between controls and postinfestation weights of dhr2-1 were significant in t-tests (both tests P < 0.001). Thus, decreased HCN release without disruption of dhurrin production resulted in increased feeding by S. frugiperda.
Figure 6.
Whole plant examination of insect (S. frugiperda) leaf consumption. Pre- and postinfestation weights were taken on whole plant BTx623 wild-type and dhr2-1 mutant individuals and compared using a one-tailed t-test. Mean weight in grams is along the x-axis and the y-axis is both pre- and postinfestation weights for the BTx623 and dhr2-1 mutant individuals.
Discussion
We have successfully demonstrated the use of next generation sequencing to identify an EMS-induced mutation responsible for cyanogenic glycoside breakdown in the complex genome of a nonmodel crop species. Previous use of NGS in targeted gene cloning has focused on model organisms [e.g., A. thaliana (Mokry et al. 2011; Uchida et al. 2011; Austin et al. 2011), Drosophila melanogaster (Blumenstiel et al. 2009; Wang et al. 2010), Caenorhabditis elegans (Sarin et al. 2008), and fission yeast (Irvine et al. 2009)]. Sorghum, while sequenced (Paterson et al. 2009), has not been used successfully to clone genes using NGS tools. To demonstrate that current NGS methods are suitable for gene cloning in a more complex genome, such as cereal crop species, we used a mutant of sorghum with a robust and easily quantifiable phenotype, HCNc.
Determining that dhr2 contained the molecular lesion by NGS was enabled by the accuracy and comprehensive view of the genome. Had we taken a more traditional positional-cloning approach, recombination would not efficiently distinguish the candidates as four dhurrinase-like paralogs, which are within 628,373 bp of each other. Substantial sequence similarity between these paralogs also complicates direct sequencing of PCR products due to coamplification of highly conserved sequences shared by multiple paralogs. The whole genome, NGS approach, however, was able to resolve these paralogs and pinpoint the exact mutation. In addition, whole genome shotgun sequencing was more cost and time efficient than targeted sequencing of the candidate gene. Currently, a single lane of Illumina HiSequation 2500 sequencing is approximately $2500 USD and library construction is $100 USD, which yields ∼37.5 Gb of data (http://www.illumina.com). Thus, a single lane provides ∼53-fold coverage of the 700-Mb sorghum genome. In this study, we obtained enough sequence data to provide a genome-wide average of 17-fold coverage. At the site of the mutation in dhr2-1 we only obtained four reads, all of which contained the G-to-A mutation. For most genes, which are not encoded by highly similar gene families, such as dhurrinases, we feel that mutation discovery could be accomplished with lower genomic coverage and therefore lower costs. At current costs for sequencing, genomic DNA libraries derived from four sorghum mutants could be sequenced to 13-fold genomic coverage for $2900 USD on a single lane at a cost per mutant of $725 USD. Comparing PCR costs for a candidate gene approach, NGS costs less than the PCR and bidirectional sequencing of a single 96-well plate of candidate gene amplicons. This NGS approach allows the discovery of SNPs in the entire genome, not just a few amplified genes. Had our mutant not been within our list of candidate genes, sequencing of the entire genome detected 576 other G-to-A mutations present in coding sequences of the mutant. Approximately half of G-to-A mutations in coding sequences result in missense, nonsense, or splice-site mutations. Annotation of the likely functions of the gene products affected by these mutations and tests of the likelihood of functional disruption by each mutation can narrow the list of candidate polymorphisms. Cosegregation of polymorphisms and the mutant phenotype can test possible causal mutations.
To expedite the process of sifting through >20 million short reads and make the method utilized widely available, we created a workflow within the Galaxy bioinformatics package (Blankenberg et al. 2001; Goecks et al. 2010; https://main.g2.bx.psu.edu/) to pinpoint the exact gene(s) responsible for the acyanogenic mutation (Figure 1). The workflow can identify and filter SNP variation from sequencing data in a semiautomated manner and rapidly identify a narrow candidate polymorphism list from anywhere in the genome. In the mutant line that contained the dhr2-1 mutation, 11,625 homozygous SNPs were identified between the reference genome of BTx623 and the acyanogenic mutant. Once homozygous SNPs were filtered for coding sequences (8.89 × 107 bp or ∼13% of the whole genome), 2131 SNPs or 18% fell within coding sequences. The SNP calling method employed by CASAVA (version 1.7.0, 2011) also provides a list of positions for which multiple reads contained the reference base and a SNP, consistent with a heterozygous polymorphic locus. As our material was substantially inbred, we hypothesize the majority of these “heterozygous” SNP positions result from sequencing or mapping errors. Consistent with the hypothesis that many of these are misalignments of repetitive sequence or the result of polymorphism between undetected paralogous sequences in the current sorghum genome assembly, only 1511 of the 14,334 heterozygous SNPs fell within coding sequences, ∼11%, a significantly lower proportion than the homozygous SNPs (P < 0.001).
The vast majority of phenotype-inducing alleles isolated from EMS-treated populations are due to G:C to A:T mutations caused by the alkylation of DNA by EMS (Henikoff et al. 2004). Among the homozygous SNPs identified in this study, only 577 mutations occurred in the coding sequences of sorghum and were G:C to A:T mutations. Among the candidate genes, a single mutation located in coding sequence was identified within the list of 577 SNPs. Thus, using knowledge about EMS mutations and a subset of the genome likely to affect our phenotype, a single G-to-A mutation in the dhr2 (dhurrinase2) gene was identified as the dhr2-1 allele. A similar approach, but using line cross linkage mapping, has been used previously to narrow the number of candidate polymorphisms down to a single causative (Schneeberger et al. 2009; Mokry et al. 2011; Uchida et al. 2011; Austin et al. 2011; Abe et al. 2012; Lindner et al. 2012; Zhu et al. 2012a,b).
Previous work has identified sorghum mutants deficient in dhurrin production and cyanide release (Blomstedt et al. 2012). The molecular lesion for one of these mutants was demonstrated to be a mutation in CYP79A1, the first enzyme in dhurrin production. Similar to the method employed in this study, identification of the mutant allele in CYP79A1 relied on previous knowledge of the dhurrin pathway. The authors first identified mutants defective in HCN release using the Feigel–Anger paper assay and then used the mutation discovery method employed in TILLING (McCallum et al. 2000a,b; Henikoff et al. 2004) to profile mutations in known biosynthetic enzymes, including CYP79A1. The authors identified a number of alleles in P450s involved in dhurrin biosynthesis as well as the glycosyl transferase, UGT85B1. In addition to the biosynthesis mutant affected in CYP79A1, the authors also identified three mutants of an unknown molecular nature, all of which were not affected in any of the P450s known to participate in the biosynthesis of dhurrin. These mutants may include alleles at dhurrinase loci, and future work will be required to determine if this is the case. Due to the ease of implementing our workflow and current sequence cost, we estimate the cost of cloning all three remaining mutants in the Blomstedt study (2012) at $2800 USD.
In this study, the identification of mutants that are not compromised in their ability to synthesize dhurrin but are affected in HCNc allowed us to characterize the effects of cyanogenesis on insect herbivory. Previously, the dhr1 (Sb08g007570) and dhr2 (Sb08g007610) genes were cloned (reviewed in Cicek and Esen 1998; Verdoucq et al. 2004; Ganjewala and Kumar 2010) and both enzymes encoded by these genes exhibit high specificity for dhurrin, their physiological substrate, as well as the structural analog sambunigrin (Hösel et al. 1987). The dhr2 gene accumulated in leaves (Thayer and Conn 1981; Hösel et al. 1987; Cicek and Esen 1998) so a mutation in dhr2 should impact herbivory and plant establishment if HCNc is an important deterrent to chewing insects. Recombinant expression of the dhurrin pathway in barley (Nielsen et al. 2006) and A. thaliana (Tattersall et al. 2001) resulted in the protection from insect predation. In Lima bean (Phaseolus lunatus L.), HCNc of 1-μM release within 10 min is enough to deter the generalist herbivore Schistocerca gregaria Forskal from feeding on leaves (Ballhorn et al. 2005). In sorghum, insect damage was negatively correlated with the levels of cyanogenic compounds and phenolic compounds (Woodhead et al. 1980). We have extended these findings to also show that S. frugiperda preferentially settled on the sorghum plants that accumulate dhurrin but have potentially very low-level release of cyanide. The dhr2-1 mutants were preferred and the insects fed to a greater degree in the absence of rapid release HCNc (Figures 3, 5, and 6). This demonstrated, using a single-gene disruption of dhr2, that sorghum cyanogenic glycosides are likely not sufficient to deter the generalist herbivore S. frugiperda in the absence of cyanide release.
Conclusions
We have successfully used NGS for forward genetics and gene cloning in sorghum. This allowed us to identify the mutation responsible for an HCNc phenotype and demonstrated that current affordable, high-volume whole genome sequencing along with available analytic methods could efficiently identify causal mutations in complex eukaryotic genomes. Isolation and characterization of the mutant allowed us to demonstrate a requirement for rapid cyanide release by dhr2 to deter insect herbivory. Like the role of cyanide release in insect deterrence, there are many biological questions that can only be answered in nonmodel organisms. The application of rapid forward genetic methods should usher in an era of fundamental insights into the genetic mechanisms responsible for biological phenomena in crops and the genetics of adaptation in any species, even those with complex eukaryotic genomes.
Supplementary Material
Acknowledgments
E.M.B. is supported by the Agriculture and Food Research Initiative Competitive grant 2012-67012-19817. C.C. was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the Department of Energy through grant DE-FG02-07ER15905. M.R.T. and K.K. were supported by The International Sorghum and Millet Collaborative Research Support Program.
Footnotes
Communicating editor: A. Charcosset
Literature Cited
- Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., et al. , 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Akazawa T., Miljanich P., Conn E. E., 1960. Studies on cyanogenic glycoside of sorghum vulgare. Plant Physiol. 35: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Amelot M. E., Oliveros-Bastidas A., 2005. Kinetics of the natural evolution of hydrogen cyanide in plants in neotropical Pteridium arachnoideum and its ecological significance. J. Chem. Ecol. 31: 315–331. [DOI] [PubMed] [Google Scholar]
- Austin R. S., Vidaurre D., Stamatiou G., Breit R., Provart N. J., et al. , 2011. Next-generation mapping of Arabidopsis genes. Plant J. 67: 715–725. [DOI] [PubMed] [Google Scholar]
- Ballhorn D. J., Lieberei R., Ganzhorn J. U., 2005. Plant cyanogenesis of Phaseolus lunatus and its relevance for herbivore–plant interaction: the importance of quantitative data. J. Chem. Ecol. 31: 1445–1473. [DOI] [PubMed] [Google Scholar]
- Blankenberg D., Kuster K. G., Coraor N., Ananda G., Lazarus R., et al. , 2001. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 19: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomstedt C. K., Gleadow R. M., O’Donnell N., Naur P., Jensen K., et al. , 2012. A combined biochemical screen and TILLING approach identifies mutations in Sorghum bicolor L. Moench resulting in acyanogenic forage production. Plant Biotechnol. J. 10: 54–66. [DOI] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson I. T., 1958. Inheritance of hydrocyanic acid production in crosses between sorghum and sudangras. Agron. J. 50: 302–306. [Google Scholar]
- Casa A. M., Pressoir G., Brown P. J., Mitchell S. E., Rooney W. L., et al. , 2008. Community Resources and Strategies for Association Mapping in Sorghum. Crop Sci. 48: 30. [Google Scholar]
- Cicek M., Esen A., 1998. Structure and expression of a Dhurrinase (β-glucosidase) from sorghum. Plant Physiol. 116: 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek M., Blanchard D., Bevan D. R., Esen A., 2000. The aglycone specificity-determining sites are different in 2, 4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA)-glucosidase (Maize beta -glucosidase) and dhurrinase (Sorghum beta -glucosidase). J. Biol. Chem. 275: 20002–20011. [DOI] [PubMed] [Google Scholar]
- Conn E. E., 1981. Cyanogenic glycosides. In: Stumpf PK, Conn EE, eds. The biochemistry of plants, pp 479–499 in A Comprehensive Treatise Secondary Plant Products, Vol. 7 Academic Press, New York. [Google Scholar]
- Conn E. E., 1994. Cyanogenesis: a personal perspective. Acta Hortic. 375: 31–43. [Google Scholar]
- De Nicola G. R., Leoni O., Malaguti L., Bernardi R., Lazzeri L., 2011. A simple analytical method for dhurrin content evaluation in cyanogenic plants for their utilization in fodder and biofumigation. J. Agric. Food Chem. 59: 8065–8069. [DOI] [PubMed] [Google Scholar]
- Dunstan W. R., Henry T. A., 1902, pp. 399–410 in Cyanogenesis in Plants. Part II. The Great Millet, Sorghum vulgare. Philosophical Transactions of the Royal Society of London. Series A, Containing Papers of a Mathematical or Physical Character, Vol. 199 Royal Society of London, London. [Google Scholar]
- Feigl F., Anger V., 1966. Replacement of benzidine by copper ethylacetoacetate and tetra base as a spot-test reagent for hydrogen cyanide and cyanogen. Analyst (Lond.) 91: 282–284. [DOI] [PubMed] [Google Scholar]
- Ferreira C., Parra J. P., Terra W. R., 1997. The effect of dietary plant glycosides on larval midgut β-glucosidases from Spodoptera frugiperda and Diatraea saccharalis. Insect Biochem. Mol. Biol. 27: 55–59. [Google Scholar]
- Ganjewala D., Kumar S., 2010. Advances in cyanogenic glycosides biosynthesis and analyses in plants: a review. Acta Biologica Szegediensis 54: 1–14. [Google Scholar]
- Glander K. E., Wright P. C., Seigler D. S., Randrianasolo V., Randrianasolo B., 1989. Consumption of cyanogenic bamboo by a newly discovered species of bamboo lemur. Am. J. Primatol. 19: 119–124. [DOI] [PubMed] [Google Scholar]
- Gleadow R. M., Woodrow I. E., 2002. Constraints on effectiveness of cyanogenic glycosides in herbivore defense. J. Chem. Ecol. 28: 1301–1313. [DOI] [PubMed] [Google Scholar]
- Goecks J., Nekrutenko A., Taylor J., 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay-Roe M., Meagher R., Nagoshi R., 2011. Effects of cyanogenic plants on fitness in two host strains of the fall armyworm (Spodoptera frugiperda). J. Chem. Ecol. 37: 1314–1322. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Till B. J., Comai L., 2004. TILLING: traditional mutagenesis meets functional genomics. Plant Physiol. 135: 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. A., 1991. The cyanogenic polymorphism in Trifolium repens L. (white clover). Heredity 66: 105–115. [Google Scholar]
- Hösel W., Tober I., Eklund S. H., Conn E. E., 1987. Characterization of β-glucosidases with high specificity for the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) moench seedlings. Arch. Biochem. Biophys. 252: 152–162. [DOI] [PubMed] [Google Scholar]
- Irvine D. V., Goto D. B., Vaughn M. W., Nakaseko Y., McCombie W. R., et al. , 2009. Mapping epigenetic mutations in fission yeast using whole-genome next-generation sequencing. Genome Res. 19: 1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. A., 1962. Selective eating of the acyanogenic form of the plant Lotus corniculatus L. by various animals. Nature 193: 1109–1110. [Google Scholar]
- Kakes P., 1991. A rapid and sensitive method to detect cyanogenesis using microtiterplates. Biochem. Syst. Ecol. 19: 519–522. [Google Scholar]
- Lindner H., Raissig M. T., Sailer C., Shimosato-Asano H., Bruggmann R., et al. , 2012. SNP-ratio mapping (SRM): identifying lethal alleles and mutations in complex genetic backgrounds by next-generation sequencing. Genetics 191: 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., et al. , 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39: D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum C. M., Comai L., Greene E. A., Henikoff S., 2000a Targeted screening for induced mutations. Nat. Biotechnol. 18: 455–457. [DOI] [PubMed] [Google Scholar]
- McCallum C. M., Comai L., Greene E. A., Henikoff S., 2000b Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123: 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V., 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokry M., Nijman I. J., Van Dijken A., Benjamins R., Heidstra R., et al. , 2011. Identification of factors required for meristem function in Arabidopsis using a novel next generation sequencing fast forward genetics approach. BMC Genomics 12: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat D. J., Clawson S., 1996. Oviposition and hatching of the clover weevil Sitona lepidus Gyll. (Coleoptera: Curculionidae). Grass Forage Sci. 51: 418–423. [Google Scholar]
- Nielsen K., Hrmova M., Nielsen J., Forslund K., Ebert S., et al. , 2006. Reconstitution of cyanogenesis in barley Hordeum vulgare L.) and its implications for resistance against the barley powdery mildew fungus. Planta 223: 1010–1023. [DOI] [PubMed] [Google Scholar]
- Oluwole O. S. A., Onabolu A. O., Link H., Roslin H., 2000. Persistence of tropical ataxic neuropathy in a Nigerian community. J. Neurol. Neurosurg. Psychiatry 69: 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., et al. , 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Sarin S., Prabhu S., Meara M. M. O., Pe I., Hobert O., 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5: 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schappert P. J., Shore J. S., 1999. Cyanogenesis, herbivory and plant defense in Turnera ulmifolia on Jamaica. Ecoscience 6: 511–520. [Google Scholar]
- Schappert P. J., Shore J. S., 2000. Cyanogenesis in Turnera ulmifolia L (Turneraceae) in developmental expression, heritability and cost of cyanogenesis. Evol. Ecol. Res. 2: 337–352. [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A. H., et al. , 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551. [DOI] [PubMed] [Google Scholar]
- Scriber J. M., 1978. Cyanogenic glycosides in Lotus corniculatus. Their effect upon growth, energy budget, and nitrogen utilization of the southern armyworm, Spodoptera eridania. Oecologia 34: 143–155. [DOI] [PubMed] [Google Scholar]
- Seigler D. S., Brinker A. M., 1993. Characterization of cyanogenicglycosides, cyanolipids, nitroglycosides, organic nitro compounds and nitrile glucosides from plants, pp. 51–131 in Methods in Plant Biochemistry, Alkaloids, Sulphur Compounds, Vol. 8, edited by Dey P. M., Harborne J. B. Academic Press, London. [Google Scholar]
- Takos A., Lai D., Mikkelsen L., Abou Hachem M., Shelton D., et al. , 2010. Genetic screening identifies cyanogenesis-deficient mutants of Lotus japonicus and reveals enzymatic specificity in hydroxynitrile glucoside metabolism. Plant Cell 22: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall D. B., Bak S., Jones P. R., Olsen C. E., Nielsen J. K., et al. , 2001. Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science 293: 1826–1828. [DOI] [PubMed] [Google Scholar]
- Thayer S. S., Conn E. E., 1981. Subcellular localization of dhurrin β-glucosidase and hydroxynitrile lyase in the mesophyll cells of sorghum leaf blades. Plant Physiol. 67: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Sakamoto T., Kurata T., Tasaka M., 2011. Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 52: 716–722. [DOI] [PubMed] [Google Scholar]
- Verdoucq L., Morinière J., Bevan D. R., Esen A., Vasella A., et al. , 2004. Structural determinants of substrate specificity in family 1 β-Glucosidases: novel insights from the crystal structure of sorghum Dhurrinase-1, a plant β-glocosidase with strict specificity, in complex with its natural substrate. J. Biol. Chem. 279: 31796–31803. [DOI] [PubMed] [Google Scholar]
- Wajant H., Mundry K.-W., 1993. Hydroxynitrile lyase from Sorghum bicolor: a glycoprotein heterotetramer. Plant Sci. 89: 127–133. [Google Scholar]
- Wang H., Chattopadhyay A., Li Z., Daines B., Li Y., et al. , 2010. Rapid identification of heterozygous mutations in Drosophila melanogaster using genomic capture sequencing. Genome Res. 20: 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R., Stiffler N., Belcher S., Kroeger T., Stern D. B., et al. , 2010. Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J. 63: 167–177. [DOI] [PubMed] [Google Scholar]
- Woodhead S., Padgham D. E., Bernays E. A., 1980. Insect feeding on different sorghum cultivars in relation to cyanide and phenolic acid content. Ann. Appl. Biol. 95: 151–157. [Google Scholar]
- Zhu Y., Mang H.-G., Sun Q., Qian J., Hipps A., et al. , 2012a Gene discovery using mutagen-induced polymorphisms and deep sequencing: application to plant disease resistance. Genetics 192: 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Smith S. M., Ayele M., Yang L., Jogi A., et al. , 2012b High-throughput discovery of mutations in tef semi-dwarfing genes by next-generation sequencing analysis. Genetics 192: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.