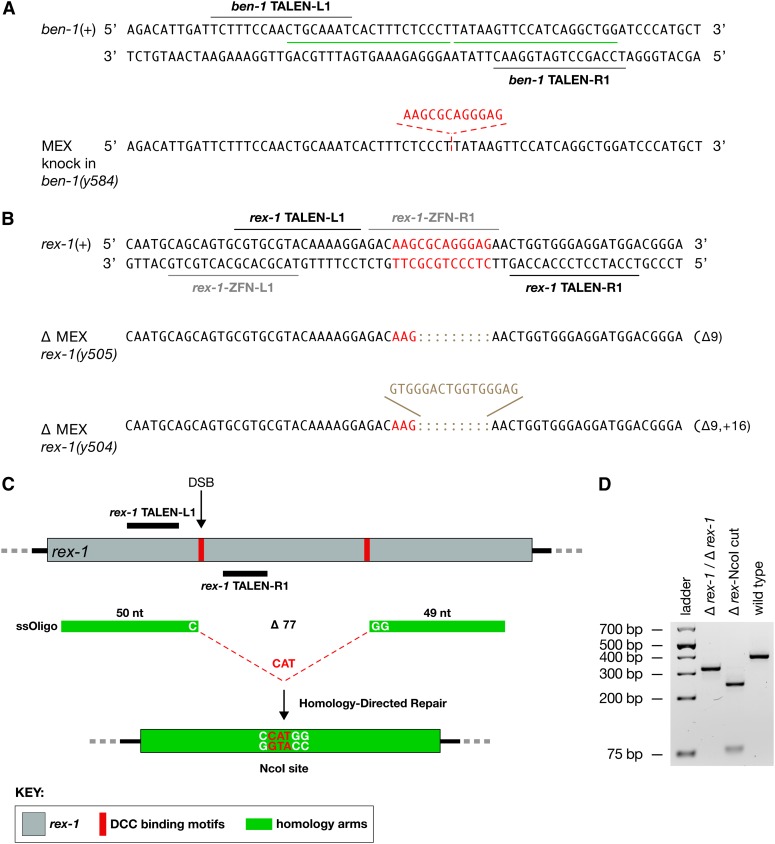
Figure 3.
Targeted HDR and NHEJ to obtain insertions and deletions of DCC binding motifs close to and relatively far from the DSB site. (A) Insertion of a DCC binding motif on an autosome. Shown is a segment of the ben-1 gene on chromosome III and the TALE recognition sequences (black lines) used to target the DSB that induced the insertion of a 12-bp DCC binding motif (MEX) by HDR. The ssOligo contained the DC binding motif (red) flanked by 20-bp homology arms (green). (B) NHEJ-mediated deletion of a DCC binding motif (MEX, in red) within rex-1. Shown is a segment of the rex-1 DCC binding site on X and the TALE recognition sequences (black lines) used to target a DSB. Sequences of representative rex-1 deletion and indel mutations generated by NHEJ are shown. Gray lines show the location of recognition sequences for ZFNs that previously caused numerous rex-1 deletions, none of which knocked out the MEX motif. (C) HDR-mediated deletion of two MEX motifs in rex-1. Schematic diagram of rex-1 (gray) shows the relative location of DCC binding motifs (red). The same TALEN pair used in B was employed to induce a DSB within the first MEX motif. The DSB was repaired by an ssOligo, including homology arms and the sequence CAT, which guided the HDR event to delete both MEX motifs. The precise deletion event was designed to remove 77 bp and insert the nucleotides CAT to create an NcoI site. (D) Agarose gel of PCR products from a rex-1 deletion strain and a wild-type strain verify the deletion. The PCR product from a precise deletion event is 326 bp. NcoI digestion of the Δ rex-1 PCR product yields a 244-bp fragment and a 82-bp fragment. The PCR product from a wild-type strain is 400 bp. The precision of the deletions was confirmed by DNA sequence.