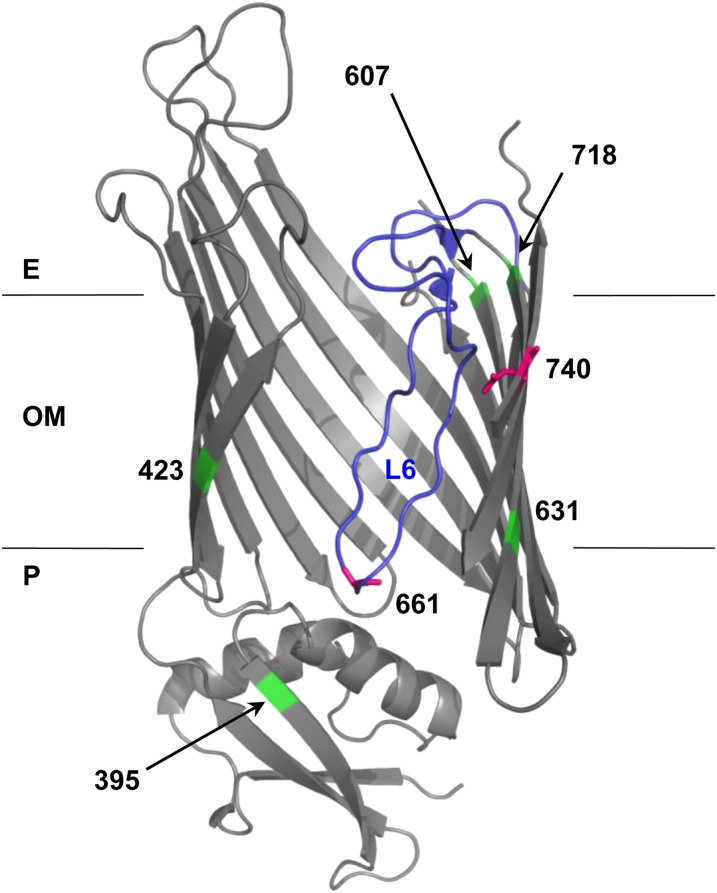
Figure 2.
Structural model of BamA mutations. BamA mutation positions (661 and 740; magenta) and suppressor positions (395, 423, 607, 631, and 718; green) were mapped onto FhaC structure 2QDZ based on the alignment of BamA and FhaC query sequences. Alignment of E. coli BamA POTRA5 (residues 347–421) and B. pertussis FhaC POTRA2 (residues 165–238) was performed using the NCBI online alignment tool COBALT (Figure S2) (Papadopoulos and Agarwala 2007). The BamA–FhaC alignment in Jacob-Dubuisson et al. (2009) was used to model β-barrel residues. Loop 6 (L6) is colored blue. Note that the loop is not well resolved in the FhaC structure, so Clantin et al. (2007) modeled it as a polyalanine chain. The outer membrane (OM), periplasm (P), and extracellular milieu (E) are indicated.