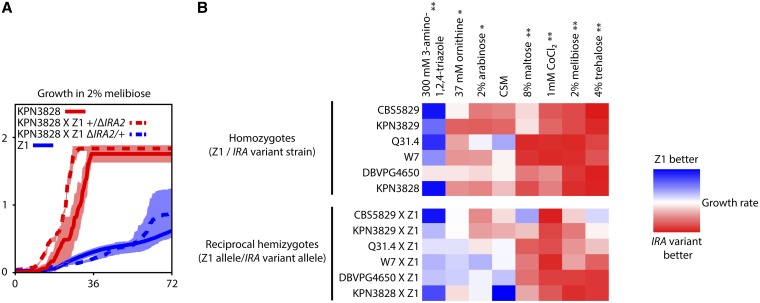
Figure 5.
Variation at IRA1 and IRA2 underlies growth rate differences in multiple conditions. (A) Shown is an example growth time course in liquid medium containing 2% melibiose as the sole carbon source, with time after inoculation on the x-axis and cell density, measured as the optical density (OD) at 600 nm, on the y-axis. Each curve reports the mean of at least two measurements of one homozygous European diploid (the flocculent strain KPN3828 or the nonflocculent strain Z1) or reciprocal hemizygote diploid constructed in the KPN3828 × Z1 hybrid background (+/ΔIRA2, bearing only the KPN3828 allele of IRA2, or ΔIRA2/+, bearing only the Z1 allele of IRA2; see Figure 3A for schematic). Error bars indicate one standard deviation. (B) Each cell reports the results of a growth experiment as in A. Color in each cell represents a ratio of the growth rates, during log-phase growth, of two strains measured in liquid culture, with each row showing data from one strain pair and each column showing data from one media condition. Top, each cell reports the ratio of growth of the homozygous nonflocculent, noninvasive European strain Z1 to growth of the indicated homozygous European strain harboring a variant IRA1 or IRA2 allele. Bottom, each row gives results from a pair of hybrid reciprocal hemizygote strains interrogating IRA1 or IRA2 in the indicated strain and Z1: each cell reports the growth of the hemizygote bearing the Z1 allele of the respective IRA gene, relative to the growth of the hemizygote bearing the variant allele at the IRA gene. Media labels at top marked by an asterisk are those in which homozygote European strains bearing variant IRA genes differed significantly from Z1 (Wilcoxon’s rank-sum test, P < 0.05 after Bonferroni correction). Media marked by ** are those inducing two significant growth effects: homozygote European strains bearing variant IRA genes differed from Z1, and hemizygote strains bearing variant IRA genes differed from hemizygotes bearing the Z1 allele at the IRA genes (Wilcoxon’s rank-sum test, P < 0.05 after Bonferroni correction). CSM, complete synthetic medium. Δ, engineered loss-of-function allele. Raw measurements and significance estimates are given in File S2.