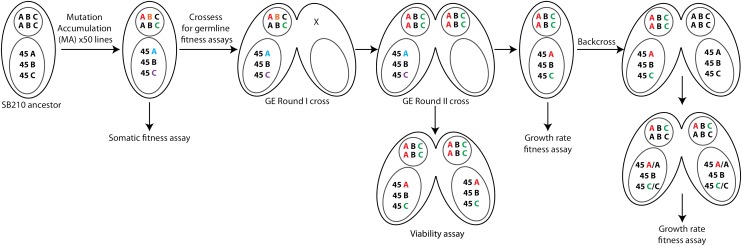
Figure 1.
Experimental design. Fifty independent MA lines were started from an SB210 ancestor. Three hypothetical loci are shown: A, B, and C. Each locus is present in two copies in the diploid germline genome and in ∼45 copies in the somatic genome. Each line was allowed to accumulate mutations independently, in both the somatic and germline genomes, shown in the lower large oval or smaller circle in each cell, respectively. Different colored letters indicate independent mutations at the three loci and two genomes. Fitness effects of mutations in the somatic genome were assayed directly on the MA lines (Somatic fitness assay). Measures of fitness effects of germline mutations required further genetic manipulations (Crosses for germline fitness assays). First, two rounds of genomic exclusion (GE) crosses were performed. In the GE round I cross, an MA line is mated with a “star” strain with a nonfunctional germline genome, indicated in the figure by an X where the germline genome should be. The resulting progeny are whole-genome homozygotes (of randomly chosen alleles) in the germline genome, but maintain the parental somatic genome. Mating in GE round II results in the formation of new somatic nuclei that are homozygous for the alleles found in the germline genome after GE round I. Mating pairs in round II GE crosses were picked and assessed for survival (Viability assay). Independently, single-cell progeny of round II GE crosses were isolated and assessed for growth rate (Growth rate fitness assay). To test for dominance effects, GE progeny were backcrossed to an ancestral cell (after GE), and heterozygous progeny were assayed for growth rate immediately after conjugation to maintain heterozygosity that would otherwise be lost to phenotypic assortment during many cell divisions.