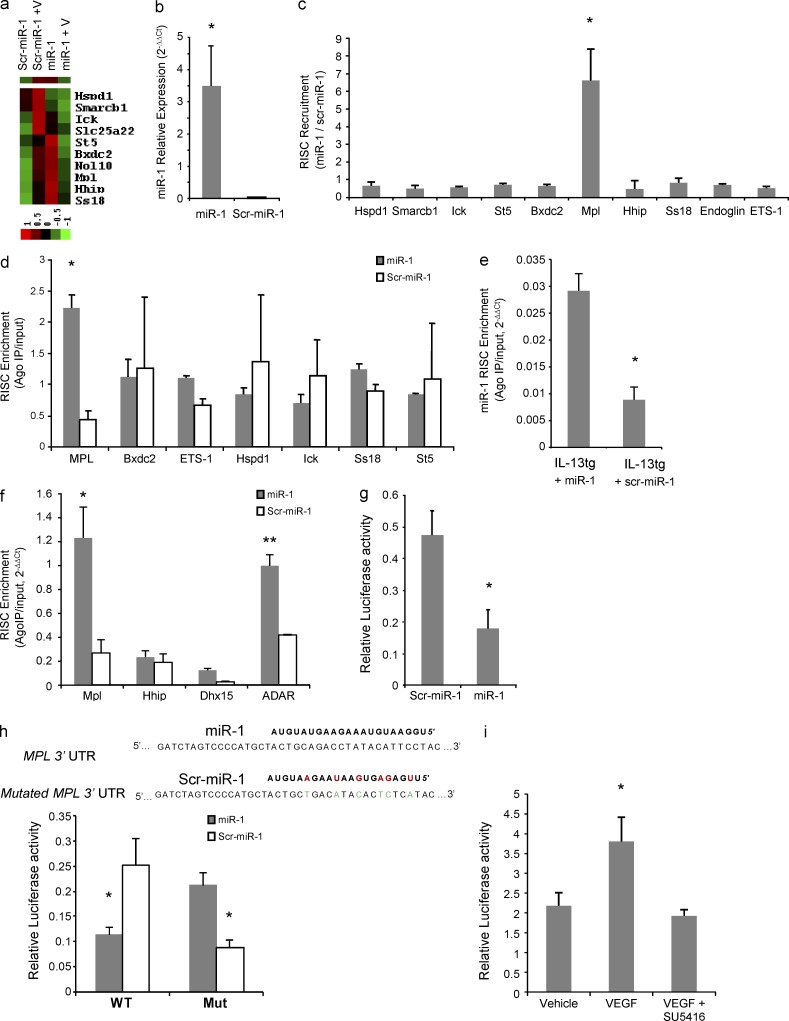
Figure 5.
Identification and validation of an miR-1 target. (a) MLECs were transfected with miR-1 or scrambled control (scr–miR-1) and stimulated with VEGF or PBS. RNA was extracted from the cells and hybridized to an mRNA microarray. Heat map shows the 10 genes that were selected as potential targets of miR-1 based on their response to VEGF stimulation and the effect of miR-1 transfection on this response. The dataset was log transformed, normalized, and median centered. The bar shows the color scale used in the heat map (values are in log base). (b–d) MLECs were transfected with miR-1 or scr–miR-1. Cells were lysed, and the lysate was immune-precipitated with anti-Ago2. (b) The level of miR-1 that coimmunoprecipitated with Ago2 was measured by TaqMan qRT-PCR and normalized to the scrambled control (data from two experiments; n = 4 in each group; *, P < 0.0001). (c) The level of mRNAs that coimmunoprecipitated with Ago2 (Ago2-IP) were measured by real-time qRT-PCR and normalized to the values for the scrambled control. The ratios represent miR-1–dependent RISC recruitment and are expressed as 2−ΔΔCt (data from three experiments; n = 4 in each group; values were compared by one-way ANOVA and corrected for multiple comparisons: *, P < 0.0002). (d) Relative concentration of each candidate mRNA in the Ago-IP fraction was divided by its relative concentration in the total cellular RNA (input fraction), and presented as 2−ΔΔCt (RISC enrichment analysis). Values for miR-1 were compared with those for scr–miR-1 (two experiments; n = 4 each group; *, P < 0.0005). (e and f) IL-13 expression was induced in IL-13 transgenic mice, and intranasal miR-1 was delivered as described in Fig. 4 c. RISC enrichment for miR-1 (*, P = 0.011) and its putative target genes (* and **, P < 0.03) were analyzed in whole lung lysates after Ago2-IP as described in c (two experiments; n = 3 per group). (g) Full-length Mpl 3′ UTR was cloned downstream from the firefly luciferase gene. This vector was cotransfected with a control Renilla luciferase vector and miR-1 or scr–miR-1, and relative luciferase activity (firefly luciferase/Renilla luciferase expression) was measured with a luminometer (three experiments; n = 5 each group; *, P < 0.0001). (h) A 56-nt fragment of Mpl 3′ UTR containing the miR-1 binding site with (Mut) or without (WT) compensatory mutations was cloned downstream of the firefly luciferase gene, and relative luciferase activity was measured as described in g (two experiments; n = 4 each; *, P < 0.005). Red letters indicate mutations in scr–miR-1, and green letters indicate compensatory mutations in Mpl 3′ UTR. (i) MLECs were transfected with firefly luciferase–Mpl3′ UTR (reporter construct) and Renilla luciferase vectors and stimulated with 100 ng/ml VEGF with or without 1 µM SU5416 (three experiments; n = 3 or more in each group; VEGF group was compared with the other two groups: *, P < 0.05). Hspd1, heat shock 60-kD protein 1 (chaperonin); Smarcb1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily b, member 1; Ick, intestinal cell (MAK-like) kinase; Slc25a22, solute carrier family 25 (mitochondrial carrier: glutamate member 22); St5, suppression of tumorigenicity 5; Bxdc2, Brix domain–containing protein 2; Nol10, nucleolar protein 10; Ss18, synovial sarcoma translocation, chromosome 18; Ets1:v-ets, erythroblastosis virus E26 oncogene homologue 1. All error bars represent mean ± SEM.