Abstract
Growth factor independence genes (Gfi1 and Gfi1b) repress recombination activating genes (Rag) transcription in developing B lymphocytes. Because all blood lineages originate from hematopoietic stem cells (HSCs) and different lineage progenitors have been shown to share transcription factor networks prior to cell fate commitment, we hypothesized that GFI family proteins may also play a role in repressing Rag transcription or a global lymphoid transcriptional program in other blood lineages. We tested the level of Rag transcription in various blood cells when Gfi1 and Gfi1b were deleted, and observed an upregulation of Rag expression in plasmacytoid dendritic cells (pDCs). Using microarray analysis, we observed that Gfi1 and Gfi1b do not regulate a lymphoid or pDC-specific transcriptional program. This study establishes a role for Gfi1 and Gfi1b in Rag regulation in a non-B lineage cell type.
Introduction
Gfi1 and Gfi1b encode 2 highly homologous nuclear proteins that function as transcriptional repressors. These proteins share a conserved C-terminal domain containing 6 zinc finger motifs that mediate DNA binding activity, and an N-terminal SNAIL/GFI-1 (SNAG) domain that mediates association with chromatin modifiers with repressive function [1-3]. Gfi1 and Gfi1b are widely expressed in the hematopoietic system [4,5]. They are both expressed in hematopoietic stem cells (HSCs) and common lymphoid progenitors (CLPs), as well as early B and T cells. Gfi1 is expressed in the monocytic and granulocytic lineages, while Gfi1b is expressed in megakaryocytic and erythrocytic lineages [6].
GFI1 and GFI1B are crucial transcriptional regulators during hematopoiesis, and play important roles in multi-lineage blood cell development [7]. Both proteins are important factors for the endothelial-to-hematopoietic transition during HSC generation, and both have been shown to restrict HSC proliferation. Gfi1 also functions to maintain self-renewal capacity and engraftment of HSCs [8]. In the myeloid compartment, Gfi1 orchestrates the linage fate decision between monocytes/macrophages and granulocytes [9]. Gfi1 deficient mice lack neutrophils, and accumulate a population of morphologically atypical immature monocytes that have the potential to generate mature macrophages but fail to produce granulocytes. Furthermore, development of dendritic cells (DCs) also depends on the expression of Gfi1, as mice lacking this protein show defective DC maturation and an overabundance of macrophages. In the lymphoid compartment, Gfi1 is important for both B and T cell development. Gfi1 deficient mice have significantly reduced numbers of B cells, and exhibit decreased thymic cellularity due to reduced proliferation, increased apoptosis and an early block at the DN stage of T cell development [10]. The exact role of Gfi1b in hematopoiesis is less well established because Gfi1b deficiency in mice results in embryonic lethality at E15 [6]. These animals likely die of failure to develop red blood cells, implicating a crucial role for Gfi1b in erythropoiesis. Gfi1b knockout mice also fail to develop megakaryocytes, but have arrested erythroid and megakaryocytic precursors in the fetal liver. In vitro, overexpression of Gfi1b inhibits myeloid differentiation of a cultured myelomonocytic cell line [11]. Recent generation of a conditional knockout model of Gfi1b has enabled analysis of the specific function of Gfi1b in adult hematopoiesis. It has been shown that B cell specific Gfi1 and Gfi1b double knockout mice have an exacerbated phenotype as compared to the Gfi1 single knockout and fail to generate any B cells [12]. This mouse model will continue to be an ideal tool to dissect the specific function of Gfi1b in different hematopoietic lineages.
Recently, we identified Gfi1 and Gfi1b as transcriptional repressors of the V(D)J recombination activating genes, Rag1 and Rag2 (collectively known as Rag), during B cell development [12]. Because Rag expression is largely lymphoid restricted, we asked whether Gfi1 and Gfi1b may play a role in repressing Rag expression in other blood lineages, which often share common transcription factor networks [13]. Furthermore, because GFI family proteins play important roles in cell fate decision during hematopoiesis, we hypothesized that they may also be responsible regulating a global lymphoid transcriptional program.
We utilized a V(D)J recombination reporter system [14] to monitor RAG activity during multi-blood lineage differentiation ex vivo when Gfi1 and Gfi1b were simultaneously deleted. We found that deletion of these genes resulted in upregulation of Rag expression in plasmacytoid dendritic cells (pDCs), but not in other blood lineages tested. However, while these Gfi1 and Gfi1b have diverse gene targets, they do not appear to regulate a lymphoid-specific transcriptional program. Our data revealed a novel role of Gfi1 and Gfi1b in Rag repression in a non-B blood lineage cell type.
Results
Deletion of Gfi1 and Gfi1b increases expression of a V(D)J recombination reporter in plasmacytoid dendritic cells in vitro
Because Gfi1 and Gfi1b repress Rag transcription in developing B cells [12], we hypothesized that they may also play a role in repressing Rag expression in non-lymphoid blood lineages that share common transcription factor networks [13]. To test this hypothesis, we utilized the H2-SVEX reporter mouse to detect RAG activity in non-B lineage cells. The H2-SVEX mouse carries a transgene expressing a violet light excited (VEX) fluorescent protein cDNA in the antisense orientation driven by a promiscuously active promoter. The cDNA is flanked by V(D)J recombination signal sequences (RSSs) oriented such that V(D)J recombination results in an inversion of the VEX cDNA into the sense orientation, irreversibly marking cells that have experienced Rag activity [14]. We generated a mouse carrying the H2-SVEX transgene and an ERT2-Cre cDNA knocked into the Rosa26 locus [15], that was also homozygous for floxed alleles of Gfi1 and Gfi1b [16,17]. The encoded ERT2-Cre protein allows for tamoxifen-inducible deletion of Gfi1 and Gfi1b.
We opted for an ex vivo system to test whether Gfi1 and Gfi1b repress Rag expression in non-lymphoid blood lineages because Gfi1 and Gfi1b deficiency results in cell lethality in multiple blood lineages in vivo [6,10,18-21]. Using established cytokine-driven culture systems, we differentiated bone marrow progenitor cells from this mouse (Gfi f/f, Gfi1b f/f, ERT2-Cre, SVEX) into macrophages, natural killer (NK) cells, megakaryocytes, conventional dendritic cells (cDCs) and plasmacytoid dendritic cells (pDCs) [22-26]. During differentiation, we treated half the culture with tamoxifen to delete Gfi1 and Gfi1b (KO), and left the other half untreated (WT). We then assayed for VEX expression in these cultures by flow cytometry. Since we observed that the background fluorescence levels differed in different culture conditions, we used a mouse of the same genotype but lacking the H2-SVEX transgene as a negative control. As expected, VEX expression was readily detected in ex vivo differentiated B cells, indicating that the reporter faithfully reflects Rag expression in culture (Figure 1A). We noted that VEX expression did not increase in progenitor B cell cultures treated with tamoxifen, suggesting that either the expected increase in Rag levels due to deletion of Gfi1 and Gfi1b is not sufficient to increase recombination, or that the recombination of H2-VEX transgene is not 100% efficient. In fact, both in our hands and in published data, only 50-85% of splenic B cells express VEX, whereas 100% of them have a history of Rag expression [27].
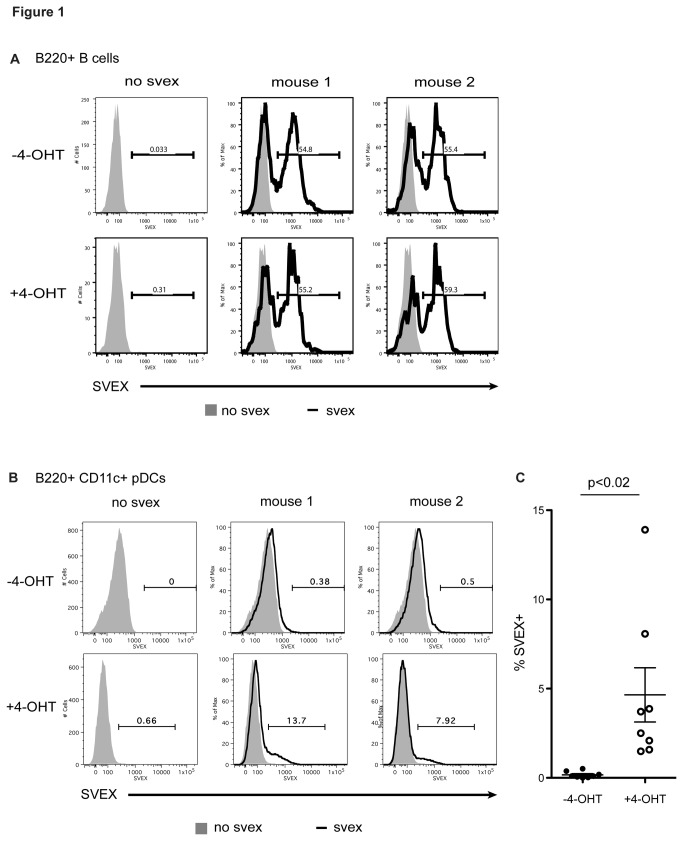
Figure 1. Deletion of Gfi1 and Gfi1b results in increased V(D)J recombination in B cells and pDCs.
(A) Flow cytometric analysis of VEX expression in B cells derived from the bone marrow of 2 individual Gfi1 f/f ; Gfi1b f/f ; ERCre, SVEX mice cultured in 5ng/ml IL-7 for 7 days (solid line), untreated (top panel) and treated (bottom panel) with tamoxifen (4-OHT). Cells were gated on B220+. Shaded histogram denotes background fluorescence from Gfi1 f/f ; Gfi1b f/f ; ERCre cells. Vertical axis ('% of max') indicates a scale of relative cell numbers with the median value set as 100%. (B) Flow cytometric analysis of VEX expression in pDCs derived from bone marrow from 2 individual Gfi1 f/f ; Gfi1b f/f ; ERCre, SVEX mice cultured in 25ng/ml Flt-3L for 8 days (solid line), untreated (top panel) and treated (bottom panel) with tamoxifen. Cells were gated on B220+ CD11c+ cells. Shaded histogram denotes background fluorescence from Gfi1 f/f ; Gfi1b f/f ; ERCre cells. Vertical axis ('% of max') indicates a scale of relative cell numbers with the median value set as 100%. All data are representative of at least three independent experiments. (C) Dot plot showing percentage of SVEX+ cells in Flt-3L cultures untreated (-4-OHT) and treated (+4-OHT) with tamoxifen. p-value was calculated using the two-tail paired Student’s t-test.
We could not detect VEX expression in ex vivo differentiated macrophages, NK cells, megakaryocytes or cDCs, either in WT and KO cultures (data not shown). In pDC cultures, however, we detected 3-15% SVEX+ cells in the tamoxifen-treated cultures but not in untreated cultures (Figure 1B). The difference in SVEX+ cells is statistically significant between cultures untreated and treated with tamoxifen (Figure 1C). These data suggest that deletion of Gfi1 and Gfi1b leads to aberrant V(D)J recombination activity in pDCs.
GFI proteins regulate Rag expression in plasmacytoid dendritic cells
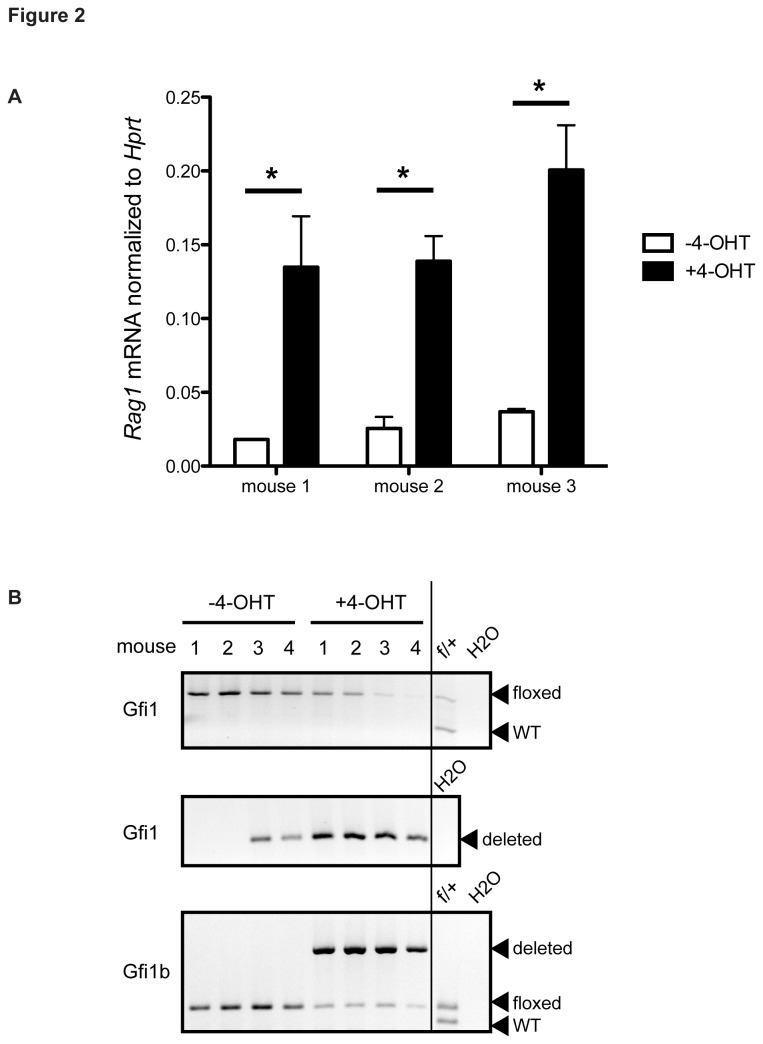
To confirm that the VEX expression in pDC cultures was indeed due to misregulated Rag expression, we sorted ex vivo differentiated pDCs using CD11c and B220 surface markers (Figure S1), and measured Rag expression by quantitative realtime PCR (RT-qPCR). As compared to pDCs derived in untreated cultures, tamoxifen-treated pDCs showed a 2-3 fold increase in Rag expression (Figure 2A), which strongly correlated with the increase in VEX expression in these cultures. The degree of de-repression was similar to that in B cells when Gfi1 and Gfi1b were deleted [12]. We observed that tamoxifen-induced deletion of Gfi1 and Gfi1b in pDC cultures was quite inefficient as assayed by genotyping PCR (Figure 2B), suggesting that the observed increase in Rag expression upon Gfi1 and Gfi1b deletion in these cultures was likely an underestimate.
Figure 2. Deletion of Gfi1 and Gfi1b results in increased expression of Rag in pDCs.
(A) Quantitative RT-PCR analysis of Rag1 transcript levels in sorted B220+ CD11c+ pDCs derived from 3 individual Gfi1 f/f ; Gfi1b f/f ; ERCre mice untreated and treated with tamoxifen (4-OHT). Values are normalized to Hprt1 transcript abundance. p-values were calculated with the two-tail Student’s t-test. * denotes p < 0.005. (B) Genotyping PCR of Gfi1 and Gfi1b loci from sorted B220+ CD11c+ pDCs derived from 4 individual Gfi1 f/f ; Gfi1b f/f ; ERCre mice untreated and treated with tamoxifen (4-OHT). Genomic DNA was isolated and subjected to PCR analysis using primers that detect wildtype (WT), floxed and deleted alleles of Gfi1 and Gfi1b. PCR products were run on 1% agarose gel and visualized with ethidium bromide. All data are representative of at least two independent experiments.
GFI proteins do not repress expression of other lymphoid genes in plasmacytoid dendritic cells
Because Rag expression is generally restricted to the lymphoid lineage, we next asked whether other lymphoid-specific genes were also regulated by GFI proteins in pDCs. We purified RNA from sorted ex vivo differentiated pDCs from untreated (WT) and tamoxifen-treated (KO) cultures and performed a microarray analysis to obtain a global view of their gene expression landscapes. We used GenePattern [28] to identify a set of genes that are differentially expressed in WT and KO pDCs by at least 2 fold (Table S1). This set of genes is not lymphoid-specific, but includes genes regulating diverse cellular processes, including cell adhesion, cytokine signaling, chemotaxis, and differentiation.
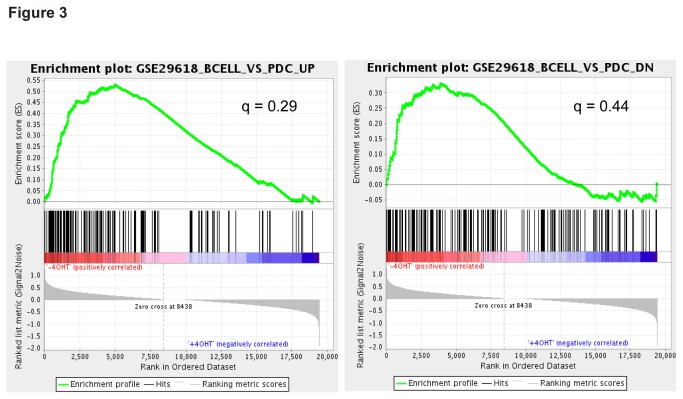
To ask whether deleting GFI proteins in pDC cultures results in a global change in lymphoid- or pDC-specific genes, we performed gene set enrichment analysis (GSEA) using curated gene sets available in the Molecular Signature Database (MSigDB) [29]. We used the GSE29618 gene sets in the C7 Immunologic Signature collection, which comprises of a set of upregulated and a set of downregulated genes when comparing gene expression profiles of B vs. pDCs [30]. We asked whether these gene sets are enriched in WT or KO pDC cultures using the false discovery rate (FDR) cutoff of 25%, a standard cutoff for GSEA analysis indicating that the probability of a false enrichment is below 25% [29]. This is represented by the q-value of < 0.25 [29]. While both gene sets showed a positive enrichment score (positively correlated with WT cultures), neither gene sets passed the FDR cutoff (Figure 3). Further, we also generated and tested gene sets that are upregulated and downregulated in B cells or pDCs when compared to HSCs using transcriptional profiling data generated by the Immunological Genome Project [31] (Table S2). We again found no statistically significant enrichment of the gene sets tested (data not shown). These results indicate that deleting GFI proteins in pDCs does not result in a global change in lymphoid- or pDC- specific genes.
Figure 3. Deletion of Gfi1 and Gfi1b does not result in misregulation of a global lymphoid or pDC program in pDCs.
Gene Set Enrichment Analysis (GSEA) was performed with the GenePattern platform. GSE29618_BCELL_VS_PDC_UP and GSE29618_BCELL_VS_PDC_DN gene sets were tested for enrichment in WT and KO pDC cultures. False discovery rate (FDR) q-values indicate the likelihood of false enrichment.
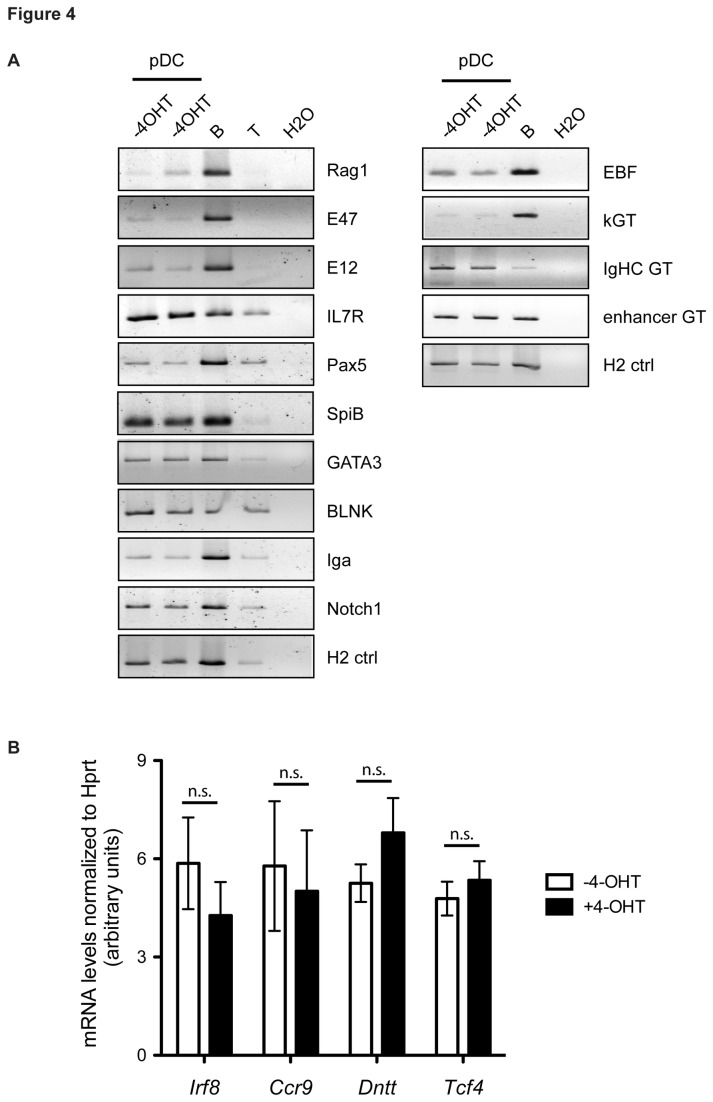
We noted that Rag and many lymphoid genes had low expression levels in WT pDCs. In fact, the list of genes found to be differentially expressed in WT vs. KO ex vivo differentiated pDCs did not include Rag because of the stringent thresholding and cutoff criteria, even though increase in Rag expression in KO cultures were confirmed by RT-qPCR before samples were subjected to microarray analysis (data not shown). To ensure that our global gene expression analysis did not miss subtle changes in the expression of individual lymphoid genes, we purified RNA from sorted ex vivo differentiated pDCs from untreated and tamoxifen-treated cultures and measured expression levels of individual genes by RT-PCR. We tested a set of lymphoid genes normally expressed in wildtype primary pDCs, including Rag [32]. We detected no increase in expression of any of the lymphoid genes tested except for Rag (Figure 4A).
Figure 4. Deleting Gfi1 and Gfi1b does not result in a change in lymphoid- or pDC-specific genes in pDCs.
(A) RT-PCR of lymphoid-specific gene expression in ex vivo differentiated B220+ CD11c+ pDCs from Gfi1 f/f ; Gfi1b f/f ; ERCre mice untreated (WT) and treated (KO) with tamoxifen. RNA isolated from primary B (B) or T (T) cells were used as controls. (B) Quantitative RT-PCR of pDC-specific genes in ex vivo differentiated B220+ CD11c+ pDCs from Gfi1 f/f ; Gfi1b f/f ; ERCre mice untreated (WT) and treated (KO) with tamoxifen. Expression values in arbitrary units are normalized to Hprt transcript abundance.
To further investigate whether GFI proteins regulate pDC-specific genes, we measured the expression of several key pDC-specific genes in WT and KO pDC cultures by quantitative RT-PCR. Two transcription factors, important for pDC development (Irf8 and Tcf4), as well as two lymphoid genes that are also highly expressed in pDCs (Dntt and Ccr9), showed no significant differences in expression in WT vs. KO pDCs (Figure 4B). Taken together, the results in Figure 4 further validate the findings of our microarray analysis, and suggest that outside the B cell lineage, GFI proteins can regulate expression of Rag, as well as a diverse set of genes, but not a global lymphoid or pDC-specific transcriptional program.
Discussion
GFI1 and GFI1B are crucial transcriptional regulators during hematopoiesis. Mouse models in which a GFP cDNA was knocked into the Gfi1 or Gfi1b loci have shown that these genes are widely expressed within the hematopoietic system [4,5]. They are essential for development of multiple blood lineages as mice deficient of Gfi1 or Gfi1b have significant defects in hematopoiesis [33]. We previously identified these proteins as repressors of Rag expression in developing B cells [12]. In this study, we demonstrated that these proteins also repress Rag expression in plasmacytoid dendritic cells (pDCs) in vitro. They, however, do not orchestrate a global lymphoid or pDC-specific transcriptional program, but regulate diverse set of genes during pDC development.
Gfi1 and Gfi1b are paralogs with very similar structures. They share conserved N-terminal and C-terminal domains, but variable intermediate region. Association of GFI1 and GFI1B with chromatin modifiers through their N-terminal SNAG domains allows them to reversibly repress their targets [3]. While it has been proposed that specific target genes may exist for GFI1 and GFI1B, both proteins share overlapping targets and exhibit functional redundancy, especially during hematopoiesis [34]. Indeed, we observed that single deletion of either Gfi1 or Gfi1b in vivo does not alter the level of Rag transcription in developing B cells, but deletion of both proteins simultaneously results in misregulation of Rag transcription in B cells [12]. In this study, we showed that deleting both Gfi1 and Gfi1b results in an increase in Rag expression in pDCs to an extent similar to that in B cells (2-3 fold). It is interesting to note that Gfi1-deficiency results in a 50% reduction in the numbers of pDC in vivo [35], implicating a role for GFI proteins in pDC development. We did not observe aberrant Rag expression in other cell types tested. However, we cannot exclude the possibility that deletion of Gfi1 and Gfi1b may affect survival of certain cell types, thus hindering the analysis of their specific function in Rag repression in these cell types.
Because Gfi1 and Gfi1b have been shown to be important for the differentiation of multiple blood lineages, we hypothesized that they may play a broader role beyond repressing Rag expression. All blood lineages originate from the hematopoietic stem cells (HSCs), which give rise to multi-potent progenitors (MPPs). These progenitors share transcription factor networks prior to commitment and restriction to a specific cell fate [36]. This phenomenon is termed transcriptional priming, and likely reflects the plasticity and the multi-lineage generation capacity of MPPs on a molecular level. Specification of cell fate thus requires the resolution of a mixed lineage gene expression pattern by induction and repression of lineage-specific genes [37-40]. Because Gfi1 and Gfi1b are crucial regulators of hematopoiesis, we postulated that they may play a role in transcriptional priming. Indeed, Gfi1 has been shown to be a direct downstream target of Ikaros, a key regulator of lymphoid priming during early hematopoiesis [41,42]. Gfi1 is part of a regulatory network that determines lineage fate decision between granulocyte and monocyte/macrophage development by antagonizing PU.1, another key factor for lineage-specific hematopoietic differentiation [42,43]. However, our microarray results suggest that these proteins play little role in specifying a lymphoid or pDC-specific transcriptional program in pDCs. While it is clear that these proteins regulate vast numbers of genes as previously shown, the gene targets are not specific to a certain lineage. Together, these data suggest that Gfi1 and Gfi1b participate in many cellular functions in pDCs, but do not regulate a lymphoid or pDC-specific gene expression profile.
Our data indicate that GFI1 and GFI1B are negative regulators of Rag in pDCs, but not in other cell types tested. Wildtype pDCs have been shown to express low levels of Rag, as well as a global lymphoid-like transcriptional program [44]. Lineage tracing experiments showed that 20-30% of pDCs have a history of Rag expression in mice [45,46]. This is believed to be an indication of the lineage affiliation of pDC development. While pDCs are clearly affiliated with the dendritic cell lineage, they show genetic and functional overlap with B cells [47]. Common lymphoid progenitors (CLPs) are capable of giving rise to pDCs [48], and pDC development is dependent on transcription factors that are also essential for B cell development, such as Ikaros, SpiB and E proteins [49-52]. Besides a set of lymphoid-specific genes including Rag, Dntt and VpreB [32], pDCs also express CD45R/B220, a B cell-specific surface marker [53]. Rag expression in pDCs is functional, as pDCs undergo partial (D-J) rearrangement at the immunoglobulin heavy chain locus [32], a hallmark of early developing B cells. Moreover, the BDCA2 receptor on pDCs has been shown to signal through signaling components of the B cell receptor, including Syk and SLP-65 [54,55]. The namesake refers to the “plasmacytoid” secretory morphology of pDCs that resembles antibody-secreting plasma B cells, and the localization and homing pattern of pDCs also resembles that of B cells [56]. These characteristics indicate that pDCs host a lymphoid-like molecular environment that is permissive to Rag expression. We set out to test the hypothesis that GFI proteins are master regulators of Rag expression, without which aberrant Rag expression would occur in all cell types. Our data, however, support a different model, where GFI proteins are acting as dampers instead of OFF-switches. This model suggests that most cell types have other robust mechanisms to suppress Rag expression, thereby preventing genomic instability. Thus, deleting Gfi1 and Gfi1b would not be predicted to alter Rag expression. However, in an environment permissive to Rag expression, such as in B cells or pDCs, GFI proteins keep Rag levels from being dangerously high. We postulate that the observed 3-15% of pDCs that aberrantly expressed Rag upon GFIs deletion were likely pDCs that already expressed Rag (20-30% of pDCs). This study demonstrates a new role for GFI proteins in regulating Rag expression in pDCs, and at the same time reveals the complex layers of regulation of Rag expression in different blood lineages.
Materials and Methods
Ethics statement
All mouse experimentation was approved by the Animal Care and Use Committee of the University of California, Berkeley (Protocol # R253-0313BR). The handling of the animals was in accordance with this protocol.
Mice
Gfi1 f/f and Gfi1b f/f mice are kindly provided by Dr. Tarik Moroy (University of Montréal). H2-SVEX mice are kindly provided by Dr. Rachel Gernstein (University of Massachusetts). ER-Cre mice were obtained from Jackson Laboratory (Bar Harbor, ME).
Chemicals
4-hydroxy-Tamoxifen was purchased from Calbiochem. Recombinant mouse IL-7 and Flt-3 ligand were purchased from R&D Systems.
Ex vivo differentiation
Total bone marrow was obtained from flushing femurs and tibias from Gfi1 f/f; Gfi1b f/f; ERCre; SVEX mice. Cells were cultured in RPMI 1640 medium supplemented with 10% (vol/vol) FCS, L-glutamine (2 mM), penicillin (100 g/ml), streptomycin (100 g/ml) and 2-mercaptoethanol (50 mM). For B cell cultures, media was supplemented with 5ng/ml IL-7. For pDC cultures, media was supplemented with 25ng/ml Flt-3L. All cells were grown at 37°C in 5% CO2. Cells were stained and analyzed by flow cytometry 7-8 days post differentiation.
Flow cytometry
Single-cell suspensions were prepared from mice or from cultured cells and were labeled with fluorochrome-conjugated antibodies by standard techniques. A FC500 (Beckman Coulter) or LSRII (BD Biosciences) flow cytometer was used for analysis; a MoFlo or an Influx high-speed cell sorter (Dako-Cytomation) was used for sorting. Data were analyzed with FlowJo software (Tree Star). Dead cells were gated out using forward and side scatter for all analyses. Analysis with ex vivo differentiated B cells was done by labeling cells with anti-B220 (RA3-6B2) antibody. Analysis with ex vivo differentiated pDCs was done by labeling cells with anti-B220 and anti-CD11c (N418) antibodies. Anti-B220 and anti-CD11b antibodies were obtained from eBioscience. Statistical significance was performed using two-tail paired Student’s t-test.
Genotyping PCR
Genomic DNA was isolated by phenol/chloroform extraction. PCR was performed with house-made Taq polymerase under cycling conditions of 95°C for 2 min, followed by 32 cycles of 95°C for 40 sec, 60°C for 40 sec and 72°C for 40 sec. Primers sequences are provided in Table S3.
Gene expression analysis by RT-PCR or quantitative real-time PCR
RNA was isolated by lysing cells in TRIzol reagent (Invitrogen). Reverse transcription was performed using MMLV-RT (Invitrogen) or SuperScript III-RT (Invitrogen) with random hexamers according to the manufacturer’s instructions. Quantitative real-time PCR was performed using JumpStart Taq polymerase (Sigma) according to the manufacturer’s protocol and fluorescent labeling with EvaGreen (Biotium). PCR cycling conditions were 95°C for 4 min followed by 40 cycles of 95 °C for 30 sec and 60 °C for 30 sec. Statistical analysis was performed using the two-tail paired Student’s t-test. RT-PCR was performed with house-made Taq with cycling condition of 95°C for 2 min followed by 32 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 40 sec. Primer sequences are provided in Table S3.
Microarray analysis
Tamoxifen-treated and untreated ex vivo differentiated pDCs from 3 independent Gfi1 f/f ; Gfi1b f/f ; ERCre mice were collected by sorting for B220 + CD11c+ cells. RNA was isolated with TRIzol reagent (Invitrogen), and further purified by RNeasy Mini kit (Qiagen). Samples were submitted for analysis to the UC, Berkeley QB3 functional genomics core facility. Affymetrix GeneChip Mouse Gene 1.0 ST Arrays (cat# 901169) were used. Differential gene expression analysis was performed using GenePattern platform (http://www.broadinstitute.org/cancer/software/genepattern/). Microarray dataset was deposited to NCBI GEO repository (GSE45837).
Bioinformatics
GSEA analysis was performed with gene sets available in the MSigDB database v4.0 curated by the GSEA Team at the Broad Institute (http://www.broadinstitute.org/gsea/msigdb/index.jsp). Analysis was performed using the GSEA module on the GenePattern platform. Custom gene sets comparing B vs. HSC and pDC vs. HSC were generated from publically available gene expression profiles of primary B, pDCs and HSCs from the ImmGen database (www.immgen.org).
Supporting Information
Gating strategy of pDC cultures. Representative gating strategy of flow cytometric analysis of pDCs derived from FLT-3L cultures. Cultures were untreated (top panel) and treated (bottom panel) with tamoxifen (4-OHT). Cells were gated on FS/SS, then CD11c+, then B220+.
(TIF)
Genes differentially expressed in WT vs. KO pDC cultures from microarray analysis.
(XLSX)
Gene lists created by comparing gene expression profiles of HSC, B and pDC, used for GSEA analysis.
(XLSX)
Primer sequences used in this study.
(DOCX)
Acknowledgments
We thank Dr. Tarik Moroy (University of Montréal) and Dr. Rachel Gernstein (University of Massachusetts) for mice, Hector Nolla and Alma Valeros (University of California, Berkeley) for help with cell sorting, and Dan Huang, Mary Stufflebeam and Jenny Chen (UC Berkeley) for assistance in mouse husbandry. We also thank all members of the Schlissel lab for experimental input and constructive criticism.
Funding Statement
This work was funded by grants to MS from NIH (RO1 HL48702 and R37 AI40227). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zweidler-Mckay PA, Grimes HL, Flubacher MM, Tsichlis PN (1996) Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol 16: 4024-4034. PubMed: 8754800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grimes HL, Chan TO, Zweidler-McKay PA, Tong B, Tsichlis PN (1996) The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol Cell Biol 16: 6263-6272. PubMed: 8887656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saleque S, Kim J, Rooke HM, Orkin SH (2007) Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell 27: 562-572. doi:10.1016/j.molcel.2007.06.039. PubMed: 17707228. [DOI] [PubMed] [Google Scholar]
- 4. Vassen L, Okayama T, Möröy T (2007) Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood 109: 2356-2364. doi:10.1182/blood-2006-06-030031. PubMed: 17095621. [DOI] [PubMed] [Google Scholar]
- 5. Yücel R, Kosan C, Heyd F, Möröy T (2004) Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol Chem 279: 40906-40917. doi:10.1074/jbc.M400808200. PubMed: 15252036. [DOI] [PubMed] [Google Scholar]
- 6. Saleque S, Cameron S, Orkin SH (2002) The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev 16: 301-306. doi:10.1101/gad.959102. PubMed: 11825872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hock H, Orkin SH (2006) Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr Opin Hematol 13: 1-6. doi:10.1097/01.moh.0000190111.85284.8f. PubMed: 16319680. [DOI] [PubMed] [Google Scholar]
- 8. Zeng H, Yücel R, Kosan C, Klein-Hitpass L, Möröy T (2004) Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J 23: 4116-4125. doi:10.1038/sj.emboj.7600419. PubMed: 15385956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dahl R, Iyer SR, Owens KS, Cuylear DD, Simon MC (2007) The transcriptional repressor GFI-1 antagonizes PU.1 activity through protein-protein interaction. J Biol Chem 282: 6473-6483. PubMed: 17197705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yücel R, Karsunky H, Klein-Hitpass L, Möröy T (2003) The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med 197: 831-844. doi:10.1084/jem.20021417. PubMed: 12682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tong B, Grimes HL, Yang TY, Bear SE, Qin Z et al. (1998) The Gfi-1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol Cell Biol 18: 2462-2473. PubMed: 9566867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schulz D, Vassen L, Chow KT, McWhirter SM, Amin RH et al. (2012) Gfi1b negatively regulates Rag expression directly and via the repression of FoxO1. J Exp Med 209: 187-199. doi:10.1084/jem.20110645. PubMed: 22201127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T et al. (2002) Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell 3: 137-147. doi:10.1016/S1534-5807(02)00201-0. PubMed: 12110174. [DOI] [PubMed] [Google Scholar]
- 14. Borghesi L, Gerstein RM (2004) Developmental separation of V(D)J recombinase expression and initiation of IgH recombination in B lineage progenitors in vivo. J Exp Med 199: 483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J et al. (2007) Restoration of p53 function leads to tumour regression in vivo. Nature 445: 661-665. doi:10.1038/nature05541. PubMed: 17251932. [DOI] [PubMed] [Google Scholar]
- 16. Zhu J, Jankovic D, Grinberg A, Guo L, Paul WE (2006) Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U S A 103: 18214-18219. doi:10.1073/pnas.0608981103. PubMed: 17116877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khandanpour C, Sharif-Askari E, Vassen L, Gaudreau MC, Zhu J et al. (2010) Evidence that growth factor independence 1b regulates dormancy and peripheral blood mobilization of hematopoietic stem cells. Blood 116: 5149-5161. doi:10.1182/blood-2010-04-280305. PubMed: 20826720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Osawa M, Yamaguchi T, Nakamura Y, Kaneko S, Onodera M et al. (2002) Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood 100: 2769-2777. doi:10.1182/blood-2002-01-0182. PubMed: 12351384. [DOI] [PubMed] [Google Scholar]
- 19. Karsunky H, Zeng H, Schmidt T, Zevnik B, Kluge R et al. (2002) Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat Genet 30: 295-300. doi:10.1038/ng831. PubMed: 11810106. [DOI] [PubMed] [Google Scholar]
- 20. Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT et al. (2003) Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18: 109-120. doi:10.1016/S1074-7613(02)00501-0. PubMed: 12530980. [DOI] [PubMed] [Google Scholar]
- 21. Horman SR, Velu CS, Chaubey A, Bourdeau T, Zhu J et al. (2009) Gfi1 integrates progenitor versus granulocytic transcriptional programming. Blood 113: 5466-5475. doi:10.1182/blood-2008-09-179747. PubMed: 19346496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shivdasani RA, Schulze H (2005) Culture, expansion, and differentiation of murine megakaryocytes. Curr Protoc Immunol Chapter 22: Unit 22F 6. PubMed: 18432953 [DOI] [PubMed] [Google Scholar]
- 23. Kouro T, Yokota T, Welner R, Kincade PW (2005) In vitro differentiation and measurement of B cell progenitor activity in culture. Curr Protoc Immunol Chapter 22: Unit 22F 2. PubMed: 18432949 [DOI] [PubMed] [Google Scholar]
- 24. Zhang X, Goncalves R, Mosser DM (2008) The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14: Unit 14 1. PubMed: 19016445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inaba K, Swiggard WJ, Steinman RM, Romani N, Schuler G et al. (2009) Isolation of dendritic cells. Curr Protoc Immunol Chapter 3: Unit 3 7. PubMed: 1965320718432791 [DOI] [PubMed] [Google Scholar]
- 26. Brawand P, Fitzpatrick DR, Greenfield BW, Brasel K, Maliszewski CR et al. (2002) Murine plasmacytoid pre-dendritic cells generated from Flt3 ligand-supplemented bone marrow cultures are immature APCs. J Immunol 169: 6711-6719. PubMed: 12471102. [DOI] [PubMed] [Google Scholar]
- 27. Borghesi L, Hsu LY, Miller JP, Anderson M, Herzenberg L et al. (2004) B lineage-specific regulation of V(D)J recombinase activity is established in common lymphoid progenitors. J Exp Med 199: 491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P et al. (2006) GenePattern 2.0. Nat Genet 38: 500-501. [DOI] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545-15550. doi:10.1073/pnas.0506580102. PubMed: 16199517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S et al. (2011) Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 12: 786-795. doi:10.1038/ni.2067. PubMed: 21743478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heng TS, Painter MW (2008) The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9: 1091-1094. doi:10.1038/ni1008-1091. PubMed: 18800157. [DOI] [PubMed] [Google Scholar]
- 32. Shigematsu H, Reizis B, Iwasaki H, Mizuno S, Hu D et al. (2004) Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity 21: 43-53. doi:10.1016/j.immuni.2004.06.011. PubMed: 15345219. [DOI] [PubMed] [Google Scholar]
- 33. van der Meer LT, Jansen JH, van der Reijden BA (2010) Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia 24: 1834-1843. doi:10.1038/leu.2010.195. PubMed: 20861919. [DOI] [PubMed] [Google Scholar]
- 34. Fiolka K, Hertzano R, Vassen L, Zeng H, Hermesh O et al. (2006) Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep 7: 326-333. doi:10.1038/sj.embor.7400618. PubMed: 16397623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rathinam C, Geffers R, Yücel R, Buer J, Welte K et al. (2005) The transcriptional repressor Gfi1 controls STAT3-dependent dendritic cell development and function. Immunity 22: 717-728. doi:10.1016/j.immuni.2005.04.007. PubMed: 15963786. [DOI] [PubMed] [Google Scholar]
- 36. Kawamoto H, Katsura Y (2009) A new paradigm for hematopoietic cell lineages: revision of the classical concept of the myeloid-lymphoid dichotomy. Trends Immunol 30: 193-200. doi:10.1016/j.it.2009.03.001. PubMed: 19356980. [DOI] [PubMed] [Google Scholar]
- 37. Yoshida T, Ng SY, Georgopoulos K (2010) Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr Opin Immunol 22: 154-160. doi:10.1016/j.coi.2010.02.011. PubMed: 20299195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ et al. (2006) Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126: 755-766. doi:10.1016/j.cell.2006.06.052. PubMed: 16923394. [DOI] [PubMed] [Google Scholar]
- 39. Lai AY, Kondo M (2006) Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med 203: 1867-1873. doi:10.1084/jem.20060697. PubMed: 16880261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Månsson R, Hultquist A, Luc S, Yang L, Anderson K et al. (2007) Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity 26: 407-419. doi:10.1016/j.immuni.2007.02.013. PubMed: 17433729. [DOI] [PubMed] [Google Scholar]
- 41. Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K (2006) Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol 7: 382-391. doi:10.1038/ni1314. PubMed: 16518393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H (2009) A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity 31: 576-586. doi:10.1016/j.immuni.2009.07.011. PubMed: 19818654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mak KS, Funnell AP, Pearson RC, Crossley M (2011) PU.1 and Haematopoietic Cell Fate: Dosage Matters. Int J Cell Biol 2011: 808524. PubMed: 21845190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sathe P, Vremec D, Wu L, Corcoran L, Shortman K (2013) Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood 121: 11-19. doi:10.1182/blood-2012-02-413336. PubMed: 23053574. [DOI] [PubMed] [Google Scholar]
- 45. Pelayo R, Hirose J, Huang J, Garrett KP, Delogu A et al. (2005) Derivation of 2 categories of plasmacytoid dendritic cells in murine bone marrow. Blood 105: 4407-4415. doi:10.1182/blood-2004-07-2529. PubMed: 15728131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Welner RS, Esplin BL, Garrett KP, Pelayo R, Luche H et al. (2009) Asynchronous RAG-1 expression during B lymphopoiesis. J Immunol 183: 7768-7777. doi:10.4049/jimmunol.0902333. PubMed: 20007571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reizis B (2010) Regulation of plasmacytoid dendritic cell development. Curr Opin Immunol 22: 206-211. doi:10.1016/j.coi.2010.01.005. PubMed: 20144853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG (2003) Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med 198: 305-313. doi:10.1084/jem.20030323. PubMed: 12874263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Allman D, Dalod M, Asselin-Paturel C, Delale T, Robbins SH et al. (2006) Ikaros is required for plasmacytoid dendritic cell differentiation. Blood 108: 4025-4034. doi:10.1182/blood-2006-03-007757. PubMed: 16912230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schotte R, Nagasawa M, Weijer K, Spits H, Blom B (2004) The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. J Exp Med 200: 1503-1509. doi:10.1084/jem.20041231. PubMed: 15583020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cisse B, Caton ML, Lehner M, Maeda T, Scheu S et al. (2008) Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135: 37-48. doi:10.1016/j.cell.2008.09.016. PubMed: 18854153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nagasawa M, Schmidlin H, Hazekamp MG, Schotte R, Blom B (2008) Development of human plasmacytoid dendritic cells depends on the combined action of the basic helix-loop-helix factor E2-2 and the Ets factor Spi-B. Eur J Immunol 38: 2389-2400. doi:10.1002/eji.200838470. PubMed: 18792017. [DOI] [PubMed] [Google Scholar]
- 53. Nakano H, Yanagita M, Gunn MD (2001) CD11c(+)B220(+)Gr-1(+) cells in mouse lymph nodes and spleen display characteristics of plasmacytoid dendritic cells. J Exp Med 194: 1171-1178. doi:10.1084/jem.194.8.1171. PubMed: 11602645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Röck J, Schneider E, Grün JR, Grützkau A, Küppers R et al. (2007) CD303 (BDCA-2) signals in plasmacytoid dendritic cells via a BCR-like signalosome involving Syk, Slp65 and PLCgamma2. Eur J Immunol 37: 3564-3575. doi:10.1002/eji.200737711. PubMed: 18022864. [DOI] [PubMed] [Google Scholar]
- 55. Cao W, Zhang L, Rosen DB, Bover L, Watanabe G et al. (2007) BDCA2/Fc epsilon RI gamma complex signals through a novel BCR-like pathway in human plasmacytoid dendritic cells. PLOS Biol 5: e248. doi:10.1371/journal.pbio.0050248. PubMed: 17850179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Randolph GJ, Ochando J, Partida-Sánchez S (2008) Migration of dendritic cell subsets and their precursors. Annu Rev Immunol 26: 293-316. doi:10.1146/annurev.immunol.26.021607.090254. PubMed: 18045026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gating strategy of pDC cultures. Representative gating strategy of flow cytometric analysis of pDCs derived from FLT-3L cultures. Cultures were untreated (top panel) and treated (bottom panel) with tamoxifen (4-OHT). Cells were gated on FS/SS, then CD11c+, then B220+.
(TIF)
Genes differentially expressed in WT vs. KO pDC cultures from microarray analysis.
(XLSX)
Gene lists created by comparing gene expression profiles of HSC, B and pDC, used for GSEA analysis.
(XLSX)
Primer sequences used in this study.
(DOCX)