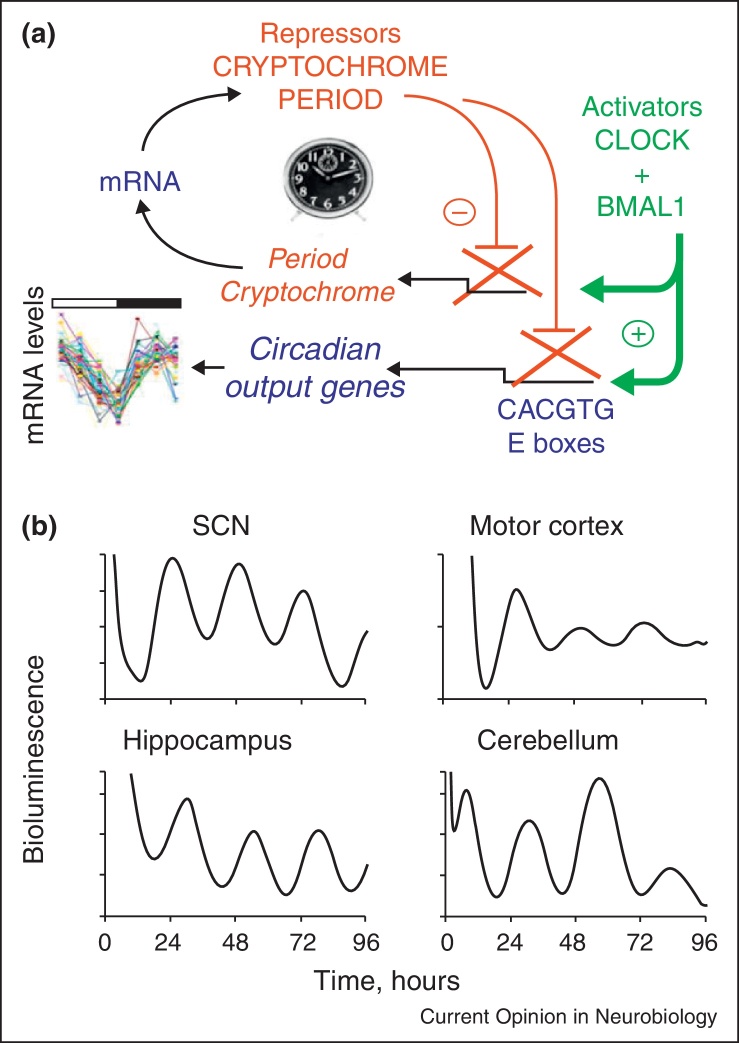
Figure 2.
The circadian clock mechanism in mammals: a negative feedback oscillator distributed widely across the brain. (a) Schematic representation of the core negative feedback loop. Heterodimeric Clock/Bmal1 complexes transactivate Period and Cryptochrome during early circadian day via E-box regulatory sequences. Following translation and nuclear localisation of their own protein products, this leads to negative regulation of these genes during early circadian night. Subsequent degradation of Per and Cry during the night releases negative regulation and transactivation can start again, defining a new circadian day. Many downstream, clock-controlled genes also carry E-boxes and so are subject to alternate phases of activation and repression across the circadian cycle, thereby directing daily cascades of cellular function. (b) Distributed circadian clocks across the mammalian brain. Bioluminescence recordings from organotypic brain slice cultures derived from mPER2:LUC reporter mice demonstrate autonomous circadian molecular cycles, not only in the principal pacemaker of the SCN, but also in other brain regions. In vivo, these distributed clocks would be synchronised to the SCN pacemaker.